Application of fusion label to promotion of Kuma030 protease soluble expression and non-affinity chromatography fast purification
A technology of fusion tag and protease, which is applied in the application field of fast protein purification method in the expression and purification of Kuma030, can solve the problems of low purification yield, low protein yield, and low-level protein expression, and achieve high purity, promote high expression, and low cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
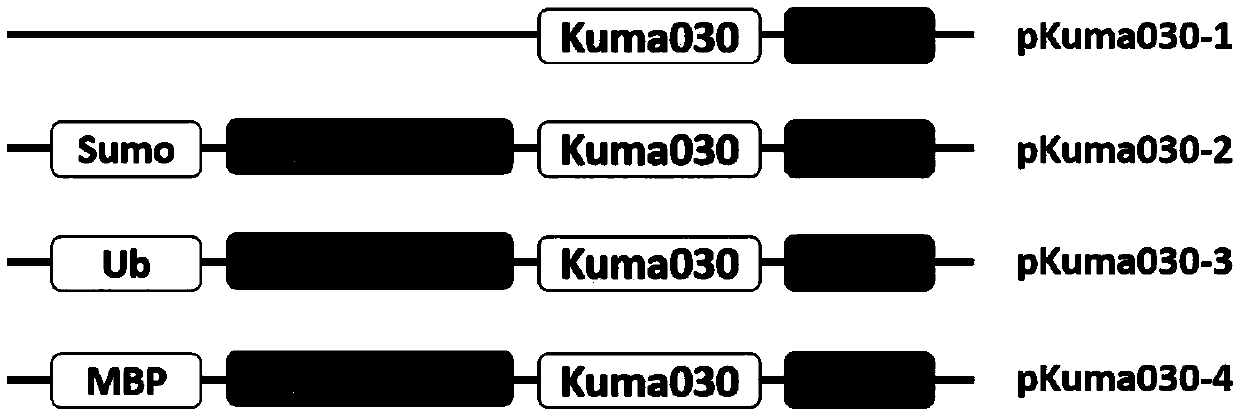
[0031] Example 1: Construction of expression vectors in which three fusion expression tags (Ub, Sumo, MBP) and Kuma030 gene are connected in series in Escherichia coli BL21 (DE3) (taking Sumo as an example)
[0032] (1) The Sumo-Kuma030 fragment was amplified by three rounds of polymerase chain reaction (PCR). The first round of PCR amplification obtained the Kuma030 gene fragment, the second round of PCR amplification obtained the Sumo fusion tag gene fragment, and the third round connected the Kuma030 gene fragment and the Sumo tag gene fragment into a large fragment (Sumo-Kuma030 ).
[0033] PCR reaction system: 10×Pfu buffer, 5 μL; dNTP (2.5mM), 5 μL; primer F (10 μM), 2 μl; primer R (10 μM), 2 μL; Pfu polymerase, 0.5 μL; Synthesis), 1 μL; add ddH2O to make the reaction system 50 μL.
[0034] PCR amplification system: 94°C, 3min; 94°C, 30s, 58°C, 20s, 72°C, 30s (extension time varies with fragment length), 25 cycles; 72°C, 5min; 4°C, ∞.
[0035] (2) After the PCR produc...
Embodiment 2
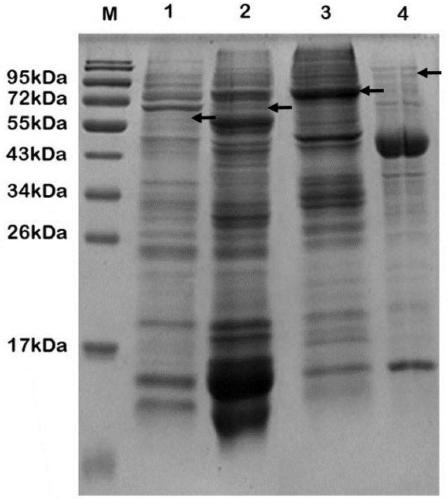
[0037] Example 2: Induced expression of Kuma030 protease containing fusion tag in Escherichia coli
[0038] The fusion expression vectors pKuma030-1, 2, 3, and 4 constructed in Example 1 were respectively transformed into Escherichia coli BL21 (DE3) competent cells. Transformation system: 1 μL of recombinant plasmid; 10 μL of competent cells; after mixing the system, place it on ice for 10 minutes, then bathe in water at 42°C for 35 seconds, add 150 μL of LB medium, incubate on a shaker at 37°C for 1 hour, and then spread it on the Incubate on plain LB plates at 37°C for 12-16h.
[0039] Inoculate a single colony in 20mL liquid LB medium, add 50μg / mL kanamycin, culture at 37°C for 12-16h, transfer to 1L liquid LB medium according to the inoculum size of 1%, add 50μg / mL kanamycin Mycin, cultured at 37°C, when OD600 reached 0.6-0.8, added IPTG with a final concentration of 0.5mM, and then transferred to 18°C for low-temperature induction for 20-24h.
[0040] After the induct...
Embodiment 3
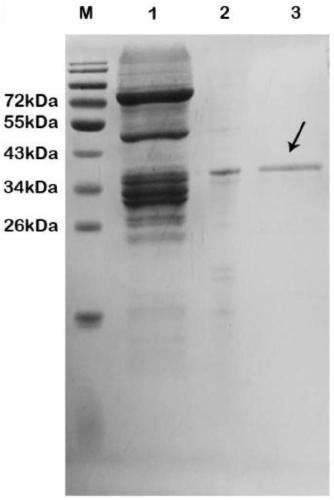
[0042] Embodiment 3: expression and Ni-NTA affinity purification of protein polypeptide substrate in Escherichia coli BL21 (DE3)
[0043] In order to verify the subsequent protease activity, the protein polypeptide substrate expression vector was constructed and expressed and purified in Escherichia coli.
[0044] (1) The fragment of MBP-substrate-GST was amplified by three rounds of polymerase chain reaction (PCR). In the first round of PCR amplification, the MBP-substrate gene fragment was obtained, in the second round of PCR amplification, the substrate-GST gene fragment was obtained, and in the third round, the MBP-substrate gene fragment and the substrate-GST gene fragment were connected into a large Fragment (MBP-substrate-GST).
[0045] PCR reaction system: 10×Pfu buffer, 5 μL; dNTP (2.5mM), 5 μL; primer F (10 μM), 2 μl; primer R (10 μM), 2 μL; Pfu polymerase, 0.5 μL; template (prk792), 1 μL; Add 50 μL of ddH2O to the reaction system.
[0046] PCR amplification syste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com