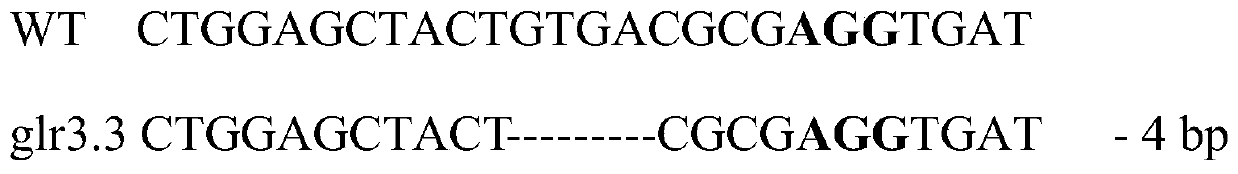Application of slglr3 Gene in Improving Plant Botrytis Botrytis Resistance
Botrytis cinerea and genetic technology, applied in the fields of application, plant peptides, plant products, etc., to achieve the effect of improving resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Preparation and identification of tomato glr3 deletion mutant plants
[0030] 1.1 Construction of CRISPR / Cas9 vector containing specific sgRNA
[0031] Find the full-length DNA sequence of GLR3 on the SGN website https: / / solgenomics.net / as shown in SEQ ID NO.3, and enter its sequence into http: / / crispr.hzau.edu.cn / cgi-bin / CRISPR2 / CRISPR website, look for the PAM sequence, that is, the sequence of 5'-NGG-3' (N represents any nucleotide of A, T, C, G), define the sequence 20 bp before NGG as sgRNA, and look for specific targeting sgRNA for the protein coding region of the gene. The DNA sequence of the sgRNA specifically targeting the protein coding region of the GLR3 gene is shown in SEQ ID NO.4.
[0032] Design CRISPR primers as follows:
[0033] CRISPR front primer (SEQ ID NO.5): CTGGAGCTACTGTGACGCGA;
[0034] Post-CRISPR primer (SEQ ID NO.6): TCGCGTCACAGTAGCTCCAG;
[0035]The intermediate vector pMD18-T was cleaved with BbsI and purified using a common...
Embodiment 2
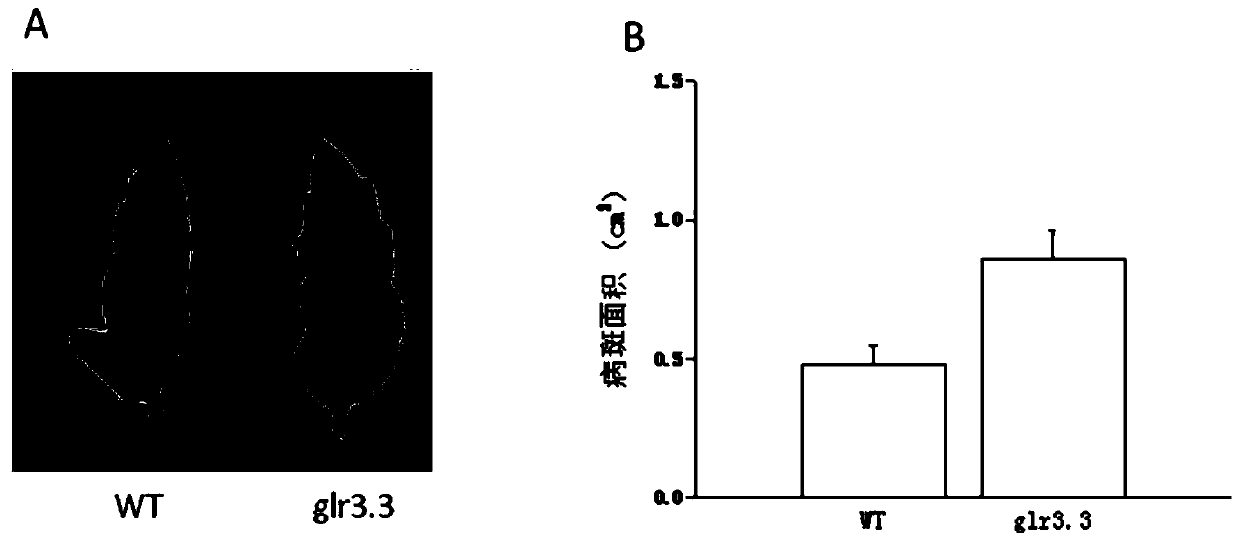
[0046] Example 2 In Vitro Identification of Tomato SlGLR3 Gene Disease Resistance Phenotype
[0047] The tomato glr3 deletion mutant leaves were inoculated with Botrytis cinerea, specifically as follows:
[0048] Botrytis cinerea pathogenic bacteria were cultivated using V8 solid medium (36% V8 fruit juice, 0.2% CaCO 3 , 2% agar powder), cultivated for about 15 days at 22°C in the dark, and after the petri dish was covered with spores, it was stored in the dark at 4°C and could be used within a week.
[0049] When in use, put the mycelia block into the inoculation medium (1% peptone, 4% maltose monohydrate), scrape off the mycelium with a tissue culture blade, vortex vigorously to release the spores, filter with gauze, and check the hemocytometer plate under the microscope Adjusted the concentration of spores down to 10 6 spores / ml is the inoculum.
[0050] Spot test on detached leaves: Take 2.5 μl of the inoculation solution with adjusted concentration, put it on the leave...
Embodiment 3
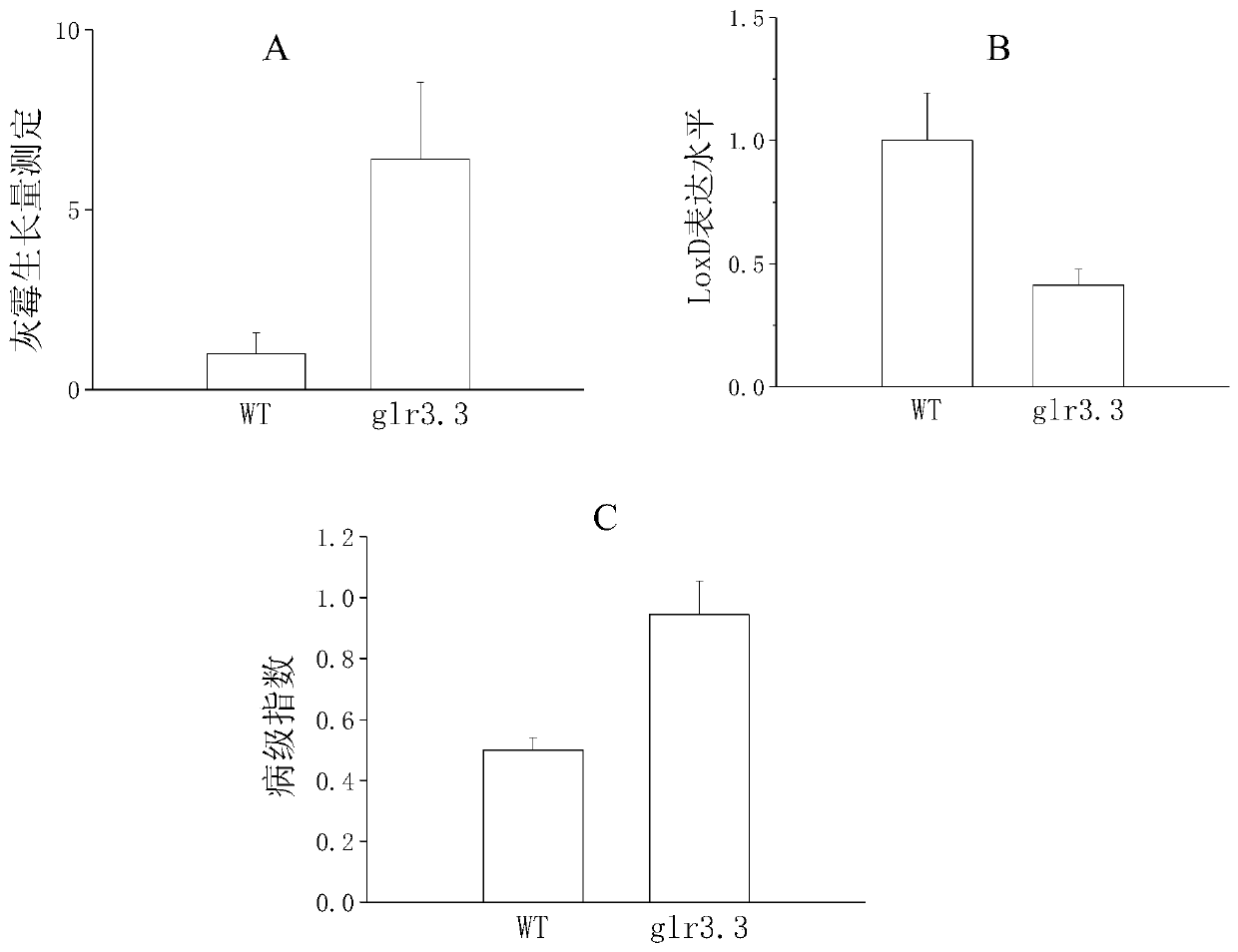
[0052] Example 3 Identification of tomato SlGLR3 gene disease resistance function and its influence on resistance hormone
[0053] The tomato glr3 deletion mutant plants were inoculated with Botrytis cinerea pathogenic bacteria, specifically as follows:
[0054] Botrytis cinerea pathogen culture V8 solid medium (36% V8 fruit juice, 0.2% CaCO 3 , 2% agar powder), cultivated for about 15 days at 22°C in the dark, and after the petri dish was covered with spores, it was stored in the dark at 4°C and could be used within a week.
[0055] When in use, put the mycelia block into the inoculation medium (1% peptone, 4% maltose monohydrate), scrape off the mycelium with a tissue culture blade, vortex vigorously to release the spores, filter with gauze, and check the hemocytometer plate under the microscope Adjusted the concentration of spores down to 10 6 spores / ml is the inoculum.
[0056] Whole plant inoculation test: Pour the adjusted inoculum solution into a 100ml watering can, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com