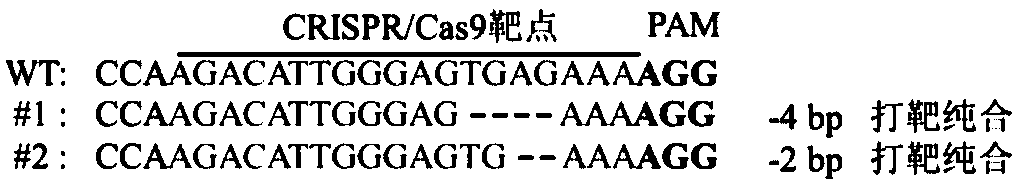Functional molecular marker of a related gene Aft for synthesis of solanum lycopersicum anthocyanin and application thereof
An anthocyanin and genotype technology, which can be used in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., and can solve problems such as co-separation of Aft sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0103] Example 1, Cloning of Tomato Aft Gene and Discovery of Allelic Variation Form thereof
[0104] 1. Analysis of the structure of the R2R3-MYB gene cluster linked to the Aft gene
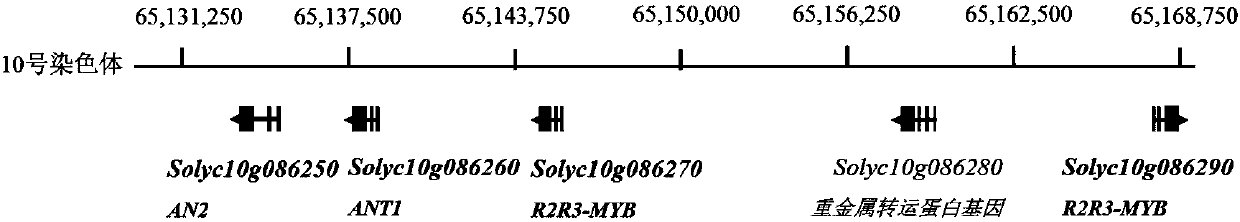
[0105] Existing studies have shown that the Aft gene is linked to a R2R3-MYB gene cluster on chromosome 10. According to tomato genome data SL2.50 version released by SGN database (URL: http: / / solgenomics.net / ), the R2R3-MYB gene cluster linked to Aft gene on tomato chromosome 10 was analyzed.
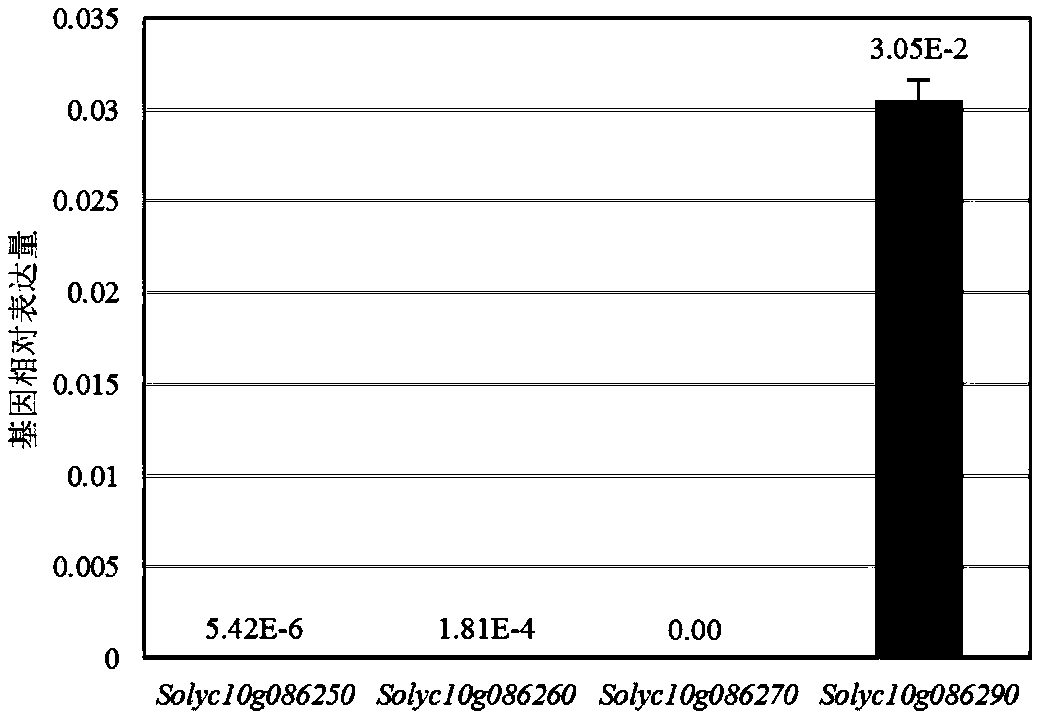
[0106] Analysis results see figure 1 . The results showed that there were four R2R3-MYB genes in the R2R3-MYB gene cluster, which were Solyc10g086250 gene, Solyc10g086260 gene, Solyc10g086270 gene and Solyc10g086290 gene. The Solyc10g086250 gene is the reported AN2 gene (Meng X, Wang J-R, Wang G-D, Liang X-Q, LiX-D, Meng QW. (2015) An R2R3-MYB gene, LeAN2, positively regulated the thermo-tolerance in transgenic tomato. J Plant Physiol 175:1-8.), Solyc10g086260 gene is the reported ANT1 gene (Mathews ...
Embodiment 2
[0125] Example 2, the development of molecular markers and polymorphism detection
[0126]1. Development of Molecular Markers
[0127] Molecular markers were developed based on the two allelic variant forms of the Aft gene.
[0128] The nucleotide sequence of the amplified molecular marker is as follows:
[0129] Primer F: 5'-GGTCACTTATTGCTGGGAGA-3' (sequence 3 in the sequence listing);
[0130] Primer R: 5'-CTCCATGTTGCATGGTTGTTG-3' (SEQ ID NO: 4 in the Sequence Listing).
[0131] 2. Polymorphism detection
[0132] A. Polymorphism detection one
[0133] The tomatoes to be tested were Indigo Rose, LA1996 or Alisa Craig.
[0134] 1. Use the Plant DNA Rapid Extraction Kit to extract the genomic DNA of the tomato to be tested.
[0135] 2. Using the genomic DNA in step 1 as a template, perform PCR amplification with the primer pair consisting of primer F and primer R in step 1 to obtain a PCR amplification product.
[0136] PCR reaction system (20 μL): 10 μL of Premix Taq DN...
Embodiment 3
[0151] Example 3, the application of molecular markers
[0152] 1. Application 1
[0153] The tomatoes to be tested were Indigo Rose, LA1996 or Alisa Craig.
[0154] 1. Carry out genotyping of the tomato to be tested according to the method of polymorphism detection one in Example 2.
[0155] 2. Plant the tomato to be tested in the field, manage it in the field regularly, and observe the phenotype of the fruit at the green ripe stage after maturity, and detect the content of total anthocyanins in the peel of the tomato to be tested.
[0156] The results showed that the pericarp of Indigo Rose had a large amount of anthocyanin accumulation, which was purple-black, and the total anthocyanin content was 7.05mg / g fresh weight; the pericarp of LA1996 had a small amount of anthocyanin accumulation, which was purple spots, and the total anthocyanin The content of Alisa Craig is 0.12mg / g fresh weight; the pericarp of Alisa Craig has no anthocyanin accumulation and is green, and the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com