Method and device for correcting high-flux sequencing data
A sequencing data, high-throughput technology, applied in sequence analysis, instrumentation, genomics, etc., can solve the problems of PCR repetition, affecting the accuracy and repeatability of test results, and low ctDNA content, so as to improve accuracy and repeatability , Improve the quality and efficiency of detection, and achieve good consistency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
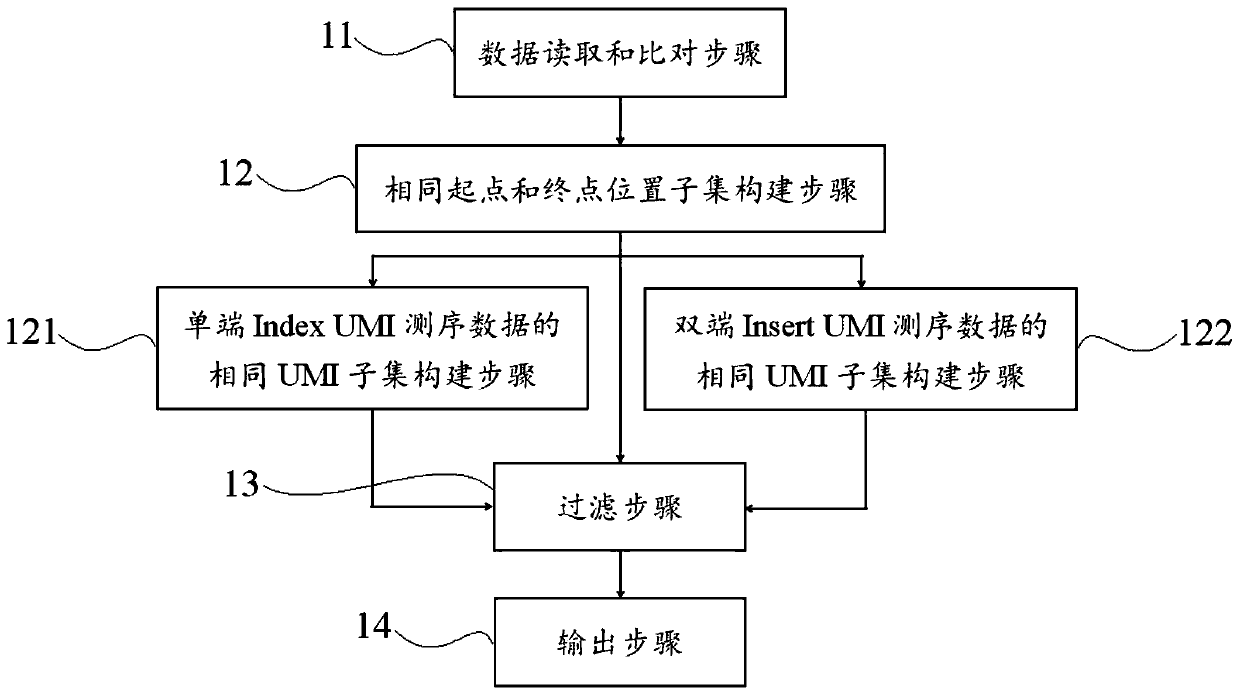
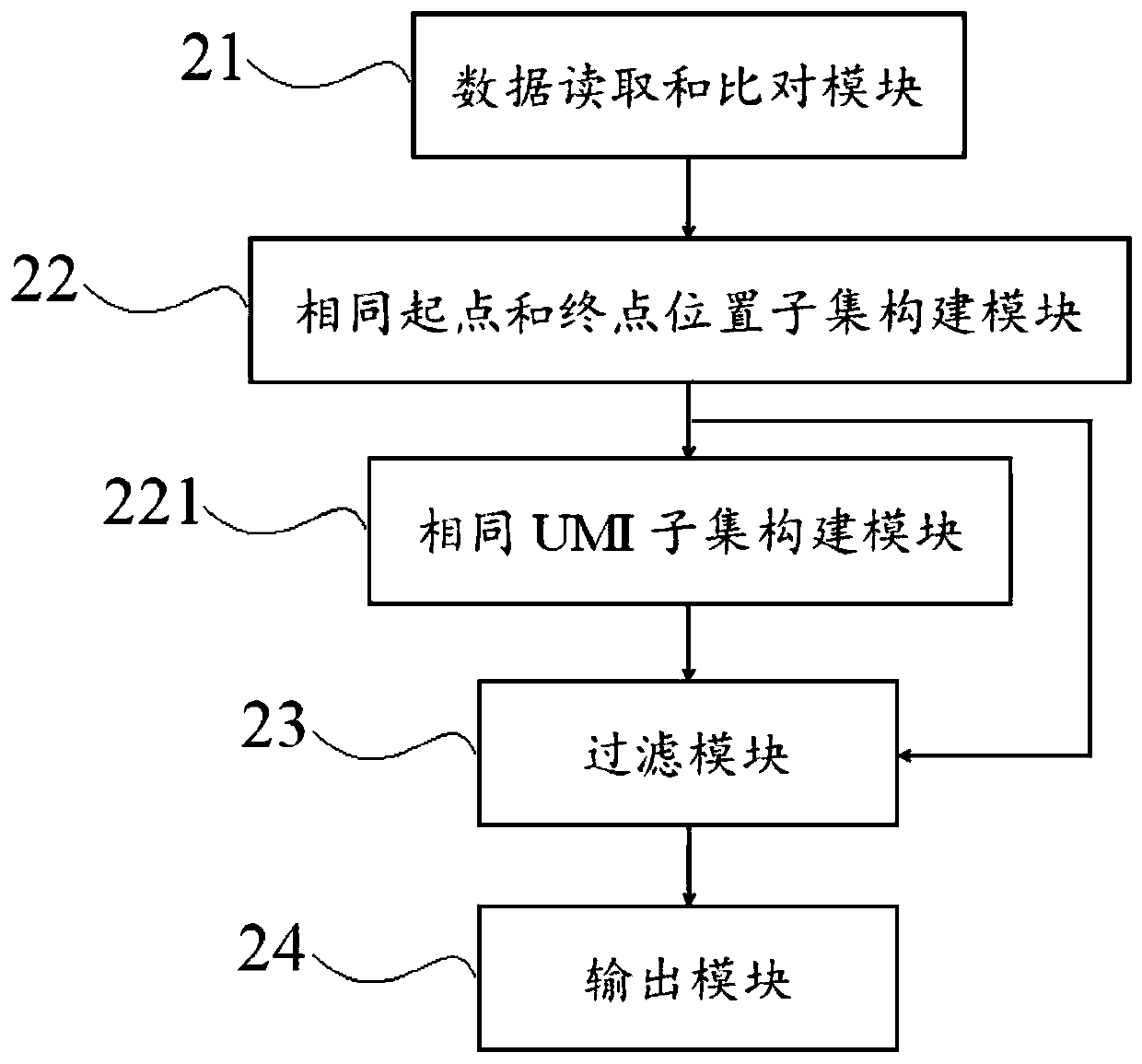
[0069] This example introduces the methods of correcting high-throughput sequencing data for different technologies in detail for non-UMI sequencing technology, single-end IndexUMI sequencing technology and paired-end InsertUMI sequencing technology, as follows:
[0070] Method 1: Calibration method for high-throughput sequencing data obtained by non-UMI sequencing technology
[0071] 1. Read the result file after alignment and sorting of the sequencing data and the reference genome, and read the reference genome sequence file at the same time.
[0072] 2. Analyze each readpair or read, and identify the chromosome, start point, and end point of the readpair or read comparison.
[0073] 3. Divide into different sets Ai according to whether the readpair or read has the same start and end positions, where i=1, 2, 3, 4, . . . . In non-UMI sequencing, the readpair or read in each Ai comes from the same DNA molecule template or the original DNA molecule double strand.
[0074] 4. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com