Kit for quantitative detection of cassava component based on micro-drop digital PCR and application thereof
A quantitative detection, droplet technology, applied in the field of molecular biology, can solve the problems affecting the accuracy of the result judgment, etc., and achieve the effect of good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Validation of specificity of cassava components
[0055] Genomic DNA of cassava, cassava, sweet potato, yam, taro, eggplant, carrot, tomato, celery, rice, soybean and barley were extracted respectively, and the extracted genomic DNA was used as a template, and the upstream primer of mSOD2 gene (tgcaagcaaagaacaaaatcgt, SEQ ID No. .1), the downstream primer of mSOD2 gene (cacaacattttccatcacaaaaaca, SEQ ID No.2) and the probe of mSOD2 gene (attaaacttctggctggtttgccccgt, SEQ ID No.3) were amplified by fluorescent quantitative PCR, and the reaction procedure of qPCR was as follows: 95°C, 5min , 1°C / s; 94°C, 15s, 1°C / s, 60°C, 1min, 1°C / s, a total of 40 cycles; 98°C, 10min, 1°C / s. Amplification curve see figure 1 , of which only cassava has a typical S-type expansion curve, and other crops have no typical expansion curve. It shows that the primer probe designed in the present invention is specific for the detection of cassava, and has good specificity.
Embodiment 2
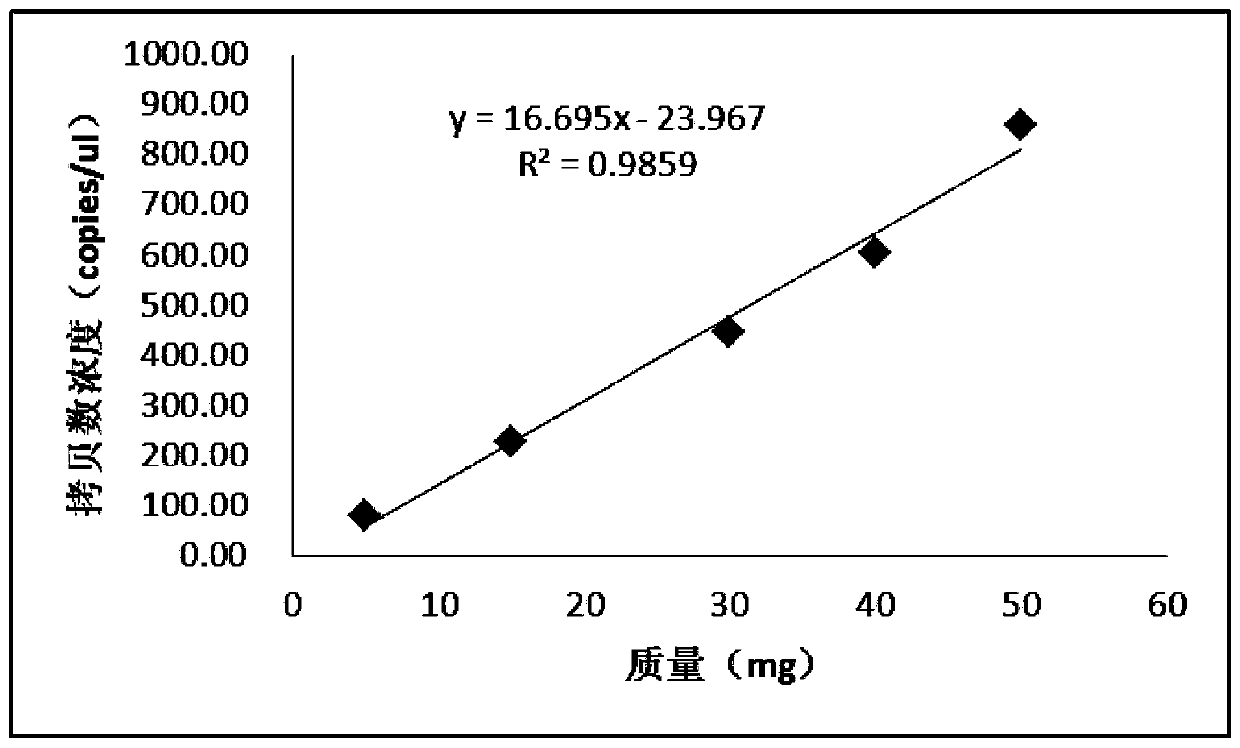
[0057] Linear relationship between cassava mass (mg) and copy number concentration (copies / μL)
[0058] Weigh 5 mg, 15 mg, 30 mg, 40 mg, and 50 mg of cassava flour samples in gradient quality, and use Wizard Genomic DNA purification kit (Promega, A1120) to extract genomic DNA, and use 20 μL digital PCR reaction system (2×ddPCRTM master mix 10 μL; primer concentration of 10 μmol / μL 0.5 μL each, 0.5 μL probe with a concentration of 10 μmol / μL, 2 μL DNA template, and water up to 20 μL. The ddPCR reaction conditions are: 95 °C, 5 min (1 °C / s); 94 °C, 15 s (1 °C / s), 60°C, 1min (1°C / s), a total of 40 cycles; 98°C, 10min (1°C / s), 12°C to store the reaction product. After the amplification, place the 96-well plate in the droplet analyzer to read Fluorescence signal, and use QuantaSoft V1.3.2 software to analyze the experimental data.
[0059] The obtained data are shown in Table 1, and the linear relationship data between cassava mass (mg) and copy number concentration (copies / μL) ar...
Embodiment 3
[0064] Determination of the Limit of Detection (LOD) and Limit of Quantification (LOQ) of Cassava DNA Concentration
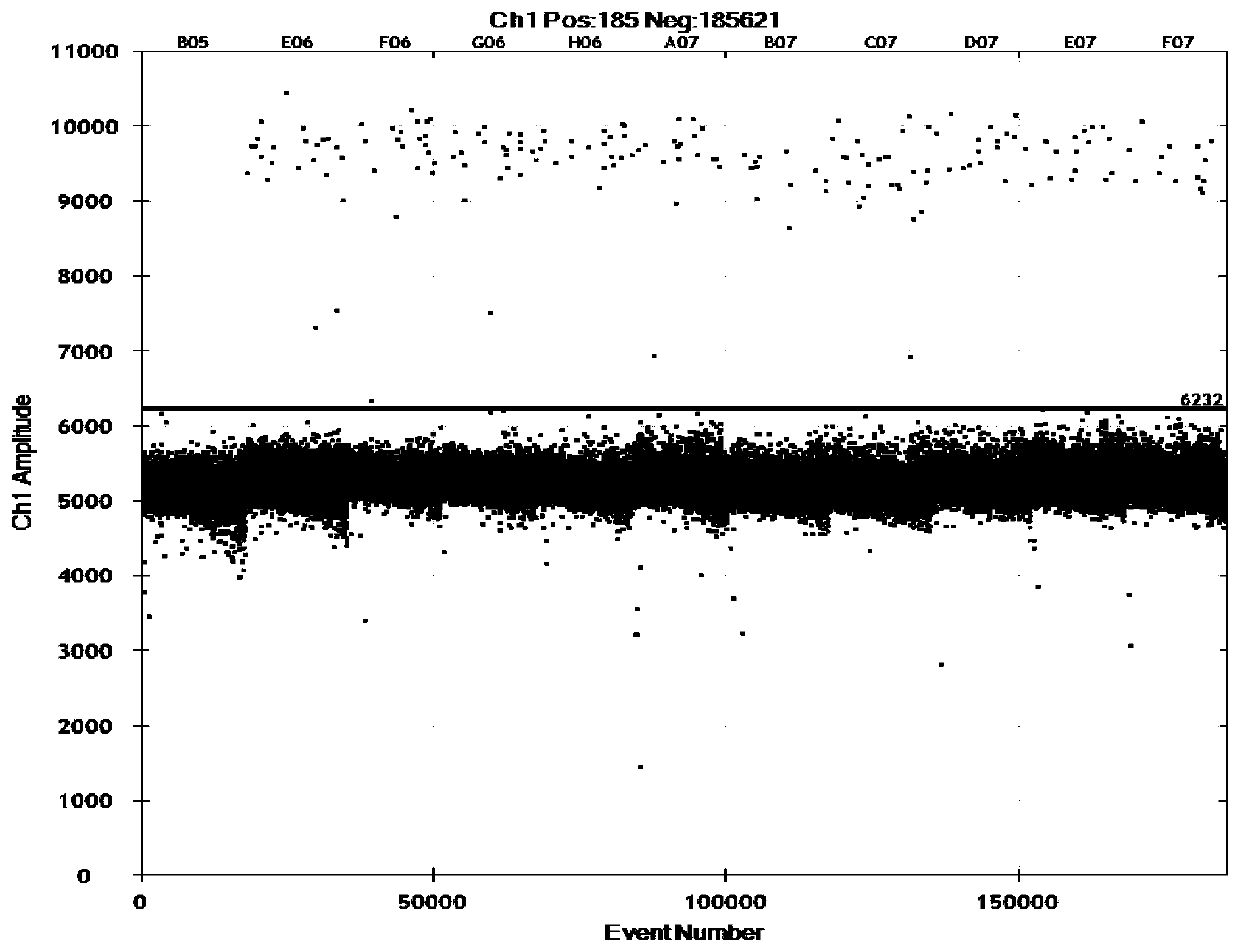
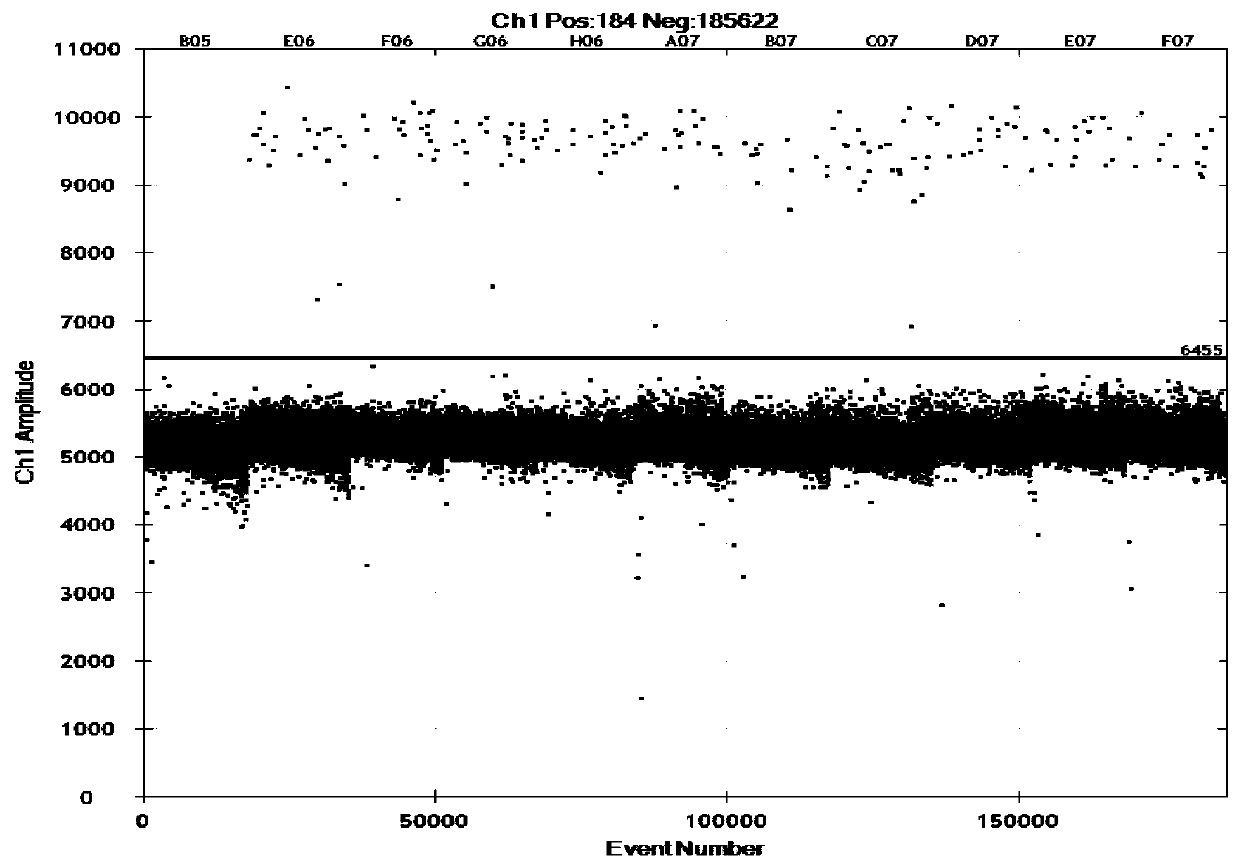
[0065] The linear relationship between the copy number concentration and DNA concentration detected by cassava-specific gene digital PCR: the DNA concentration gradients were 18ng / μL, 3.6ng / μL, 1.8ng / μL, 0.6ng / μL and 0.2ng / μL, and each gradient was three parallel. Judgment conditions are: LOQ is the lowest copy number concentration corresponding to the relative standard deviation (RSD) of the detection result being less than 25%, and LOD is the lowest copy number concentration that can be detected by no less than 9 out of 10 parallels. The data of cassava DNA concentration copy number test result are shown in Table 2. The heat map of cassava DNA concentration copy number test is shown in image 3 . The heat map of cassava DNA concentration gradient LOD and LOQ test is shown in Figure 4.
[0066] Table 2 cassava DNA concentration copy number test results
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com