Blood exosome molecular marker and application thereof in preparation of liver cancer diagnostic products
A technology of molecular markers and exosomes, applied in the field of biomedicine, can solve problems such as relying on tissue biopsy, achieve convenient non-invasive diagnosis methods, improve sensitivity and specificity, and be easy to detect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Preparation of exosomes
[0060] When the liver cancer cell lines Huh7 and HepG2, and the normal liver cell line HL-7702 were conventionally cultured to a density of about 40%, they were washed twice with PBS and then cultured in a culture system supplemented with exosome-free serum. After 48 hours, the cell supernatant was collected, and the exosomes were separated and prepared by ultracentrifugation. Centrifuge at 300g at 4°C for 10 minutes; take the supernatant and centrifuge again (4°C, 2000g, 10 minutes); take the supernatant and centrifuge again (4°C, 10,000g, 30 minutes). The precipitate was discarded, and the supernatant was centrifuged at 100,000g for 70 minutes; the collected precipitate was resuspended in PBS, and then centrifuged again (100,000g, 70 minutes), and the obtained precipitate was cell exosomes.
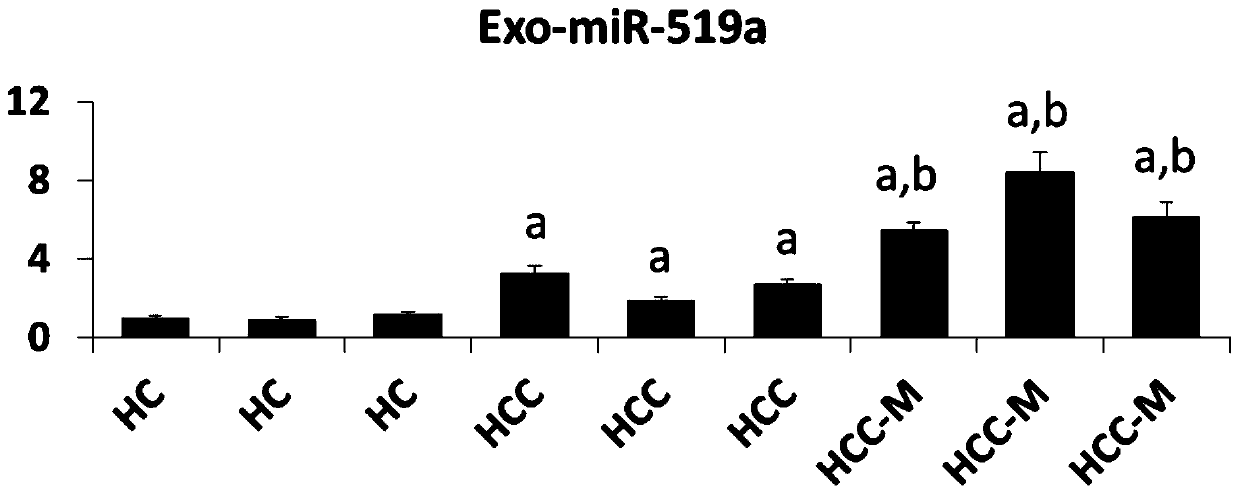
[0061] Collect anticoagulant and non-anticoagulant blood from patients or medical examiners, centrifuge at 2000 rpm for 10 minutes at 4°C, se...
Embodiment 2
[0062] Example 2 Identification of exosomes
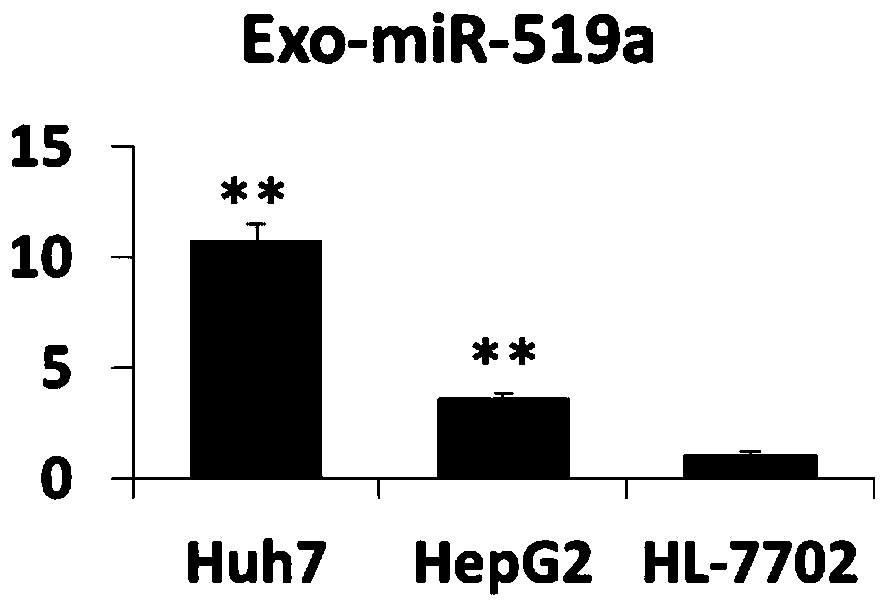
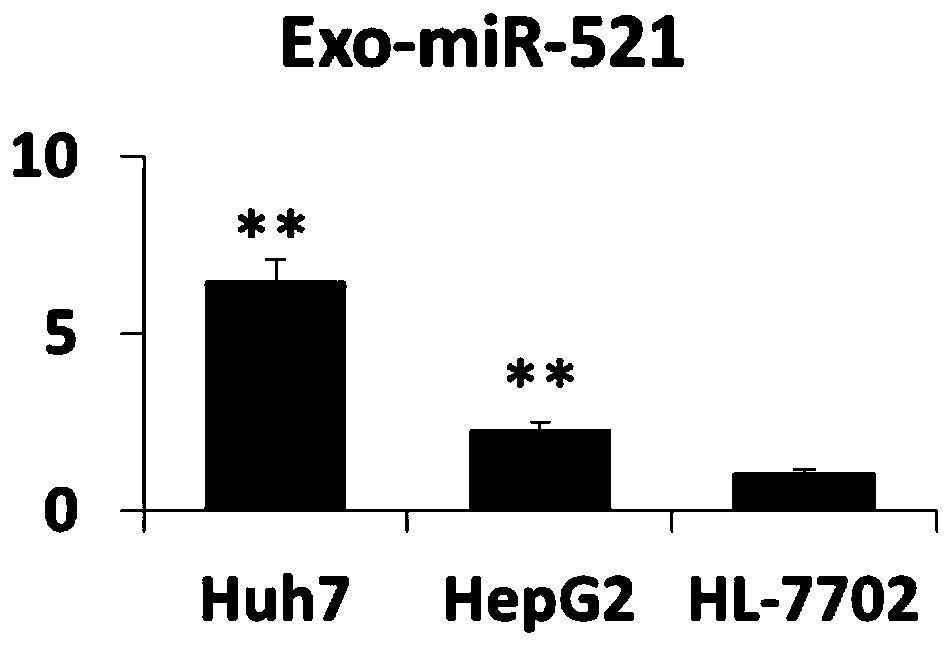
[0063] The particle size of cell supernatant and plasma or serum exosomes was analyzed by NTA method (Nanoparticle Tracking Analysis: nanoparticle tracking analysis method). The particle size range is 35-135nm. The exosome markers on the surface were detected by Western Blot or magnetic bead method (flow cytometry). Western Blot detection showed that the isolated Huh7 cells and the plasma and serum exosomes of patients with liver cancer all expressed CD63, CD9 and CD81.
Embodiment 3
[0064] Example 3 Analysis of the expression level of exosome miR-519a
[0065] Collect exosomes from Huh7, HepG2, and HL-7702 cells, or exosomes from liver cancer patients and healthy controls, resuspend them in 200 μL of PBS, place them on ice, and add 200 μL of 2X Denaturing Solution from Thermo Fisher to mix. and then add 25 fmol of external reference Cel-miR-39. Then miRVana miRNA extraction kit from Thermo Fisher Company was used to separate and prepare miRNA. Then, the miRNA reverse transcription kit was used, and the reverse transcription primer of miR-519a (CAGCATAGGTCACGCTTATGGAGCCTGGGACGTGACCTATGCTGCAGAAA) was used for reverse transcription. The reaction program was: 15 minutes at 42°C, 5 seconds at 85°C. The PCR detection primer pair of miR-519a was then used for fluorescence quantitative PCR detection. The PCR system was: 2×SYBR Green Mix-10 μl; miR-519a upstream primer (GCCGGGCTCTAGAGGGAAGCGC) 5 μM-0.8 μl; miR-519a downstream universal primer ( GTCACGCTTATGGAGCC...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com