Nucleic acid molecules, expression vectors and applications, antibacterial drugs and methods
A nucleic acid molecule and expression vector technology, which is used in antibacterial drugs, nucleic acid molecules, expression vectors and application fields, and can solve problems such as rare reports on histone genes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
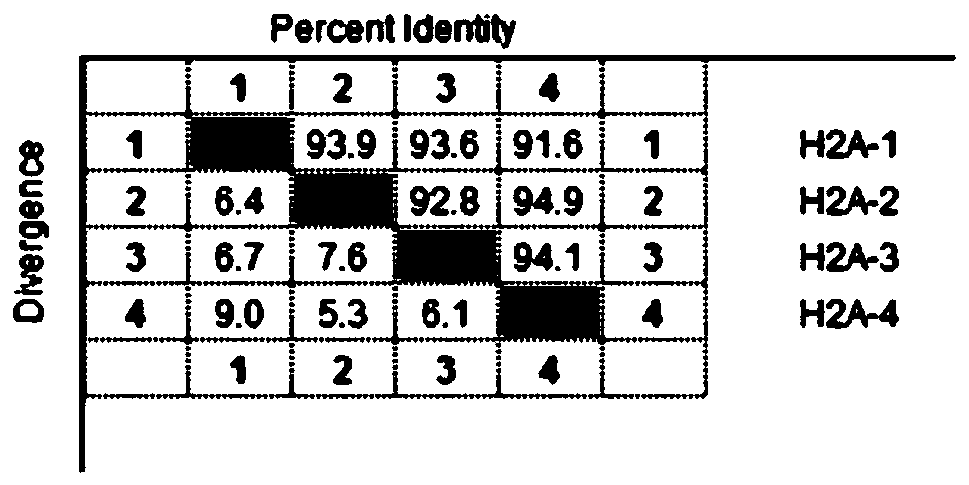
[0040] In the process of studying the histone H2A of zebrafish, the inventors found that there are abundant nucleotide polymorphisms in the gene of the protein. The inventors further isolated four variant nucleic acid sequences of histone H2A, and named them H2A-1, H2A-2, H2A-3, and H2A-4, respectively. Sequence comparative analysis shows that the sequences of the four variant nucleic acid molecules are respectively shown in SEQ ID NO.1-4. There are a total of 42 base differences in these 4 variant nucleic acid sequences ( figure 1 ). The identity between the nucleic acid sequences of these four variants is between 91.6% and 94.9% ( figure 2 ). Among them, the amino acid sequences of the histones encoded by H2A-1, H2A-3, and H2A-4 are consistent, as shown in SEQ ID NO.5, but the amino acid sequences of these three are the same as those of the histones encoded by H2A-2 (such as SEQ ID NO.6) sequence is inconsistent, reflected in the 17th and 42nd amino acid residue types a...
experiment example 1
[0043] Effect of overexpression of the above-mentioned variant nucleic acid sequences on the proliferation of Edwardsiella caturis
[0044] In this experiment example, the influence of the overexpression of H2A-1, H2A-2, H2A-3 and H2A-4 in fish on the proliferation of Edwardsiella caturis was tested by the plate counting method. The specific operations are as follows:
[0045] 1.1 By microinjection, empty plasmids and plasmids that can express H2A-1, H2A-2, H2A-3 and H2A-4 respectively (see Figure 7 ) were microinjected into zebrafish embryos respectively;
[0046] Insert the complete open reading frames of H2A-1, H2A-2, H2A-3 and H2A-4 into the eukaryotic expression vector p3XFLAG-CMV TM -14 in.
[0047] 1.2 On the 4th day after fertilization, after the zebrafish larvae hatched, the empty plasmid group, H2A-1, H2A-2, H2A-3 and H2A-4 plasmid groups were divided into groups for soaking infection, catfish Edwards Bacterial infection concentration is 2×10 8 pfu / ml;
[0048]...
experiment example 2
[0053] Effects of in vivo overexpression of the above-mentioned variant nucleic acid sequences on the survival rate of fish infected with Edwardsiella caturis
[0054] In this experiment example, the influence of the overexpression of the three variant nucleic acid molecules of H2A-1, H2A-2 and H2A-3 in zebrafish on the survival rate under the infection of Edwardsiella catguis was tested by infection survival curve. The specific operation is as follows:
[0055] 1.1 Microinject empty plasmid FLAG, plasmids expressing H2A-1, H2A-2 and H2A-3 into zebrafish embryos by microinjection;
[0056] 1.2 On the 4th day after fertilization, after the zebrafish larvae hatched, the empty plasmid group, H2A-1, H2A-2 and H2A-3 plasmid groups were divided into groups for soaking infection, and the infection concentration was 2×10 8 pfu / ml;
[0057] 1.3 Count the number of surviving juveniles every day for 6 days. The survival rate curve was drawn by GraphPad Prism software.
[0058] Experi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com