Preparation method of multi-primer oligo probes
A multi-primer and probe technology, which is applied in the field of preparation of multi-primer oligo probes, can solve the problems of inability to disassemble the probe and limit the application of oligo-FISH technology, and achieve the effect of improving the application ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
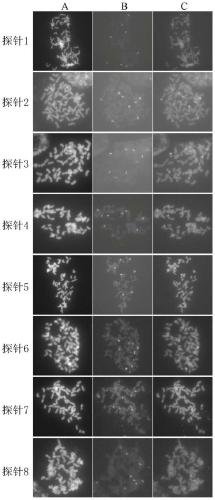
[0018] Embodiment 1: the preparation method of multi-primer oligo probe
[0019] 1. Preliminary design of the oligo sequence: based on the sequenced Saccharum genus SES208 genome, first use the RepeatMasker software to filter out the repetitive sequence of the genome and design the remaining sequence as a 59bp oligo sequence, compare the sequence back to the genome to remove multiple Align the oligo sequences at the sites (homology > 75%), and use Primer5 to calculate the Tm value and hairpin Tm value of each remaining oligo sequence, and those with a difference > 10°C are retained.
[0020] 2. Add primers at both ends of the oligo sequence: take a 2mb interval on Chromosome 1-8 as an independent probe, and add the same primer sequence F1 to the 5' end of the oligo sequence of each independent probe, Add different primer sequences R1-R8 to the 3' ends of the oligo sequences of different probes, and the oligo sequences of 8 intervals form an oligo sequence library and send it t...
Embodiment 2
[0022] Example 2: Multi-primer oligo-FISH technology
[0023] The multi-primer oligo-FISH technology refers to the addition of different primer sequences R1-R8 to the 3' end of the oligo sequence of different probes when designing the oligo sequence for the preparation and differentiation of probes and then combined with fluorescence in situ hybridization technology. The method can split an oligo probe sequence library into 8 small probes to improve the application ability of FISH technology in more precise molecular cytogenetics research
[0024] The application steps of multi-primer oligo-FISH of the present invention are:
[0025] 1. Preparation of metaphase chromosomes: take the young root tips of SES208 and put them into 2mM 8-hydroxyquinoline solution for two hours to pretreat to enrich metaphase cells; then fix the root tips with ethanol: acetic acid (3:1) fixative solution, The root cap and elongation zone were removed, and the meristematic zone was hydrolyzed in a mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com