Computer-aided screening method for small molecule target nucleic acid aptamers based on high-performance computing platform and small molecule target nucleic acid aptamers
A high-performance computing and computer-aided technology, applied in the field of molecular biology, which can solve the problems of nucleic acid molecule grabbing target, difficult separation, loss of target sequence, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1: If the energy of the target small molecule target is -5Kcal / mol when it binds to the conformation of nucleic acid a, and the energy when it binds to the b conformation is -6Kcal / mol, then it is more stable when it binds to the b conformation than when it binds to the a conformation. This value is generally Below -3Kcal / mol is considered desirable, therefore, in general, the b conformation is desirable.
Embodiment 2
[0074] Example 2: For example, when the target small molecule target binds to the conformation of nucleic acid a, the energy is -5Kcal / mol, and when it binds to the b conformation, the energy is -6Kcal / mol, but the main hydrophobic part in the b conformation is bound to the affinity of the target small molecule target. At the water end, even if the energy of the b conformation is low, the b conformation is not advisable.
Embodiment 3
[0075] Example 3: For example, when the target small molecule target binds to the conformation of nucleic acid a, the energy is -5Kcal / mol, and when it binds to the b conformation, the energy is -6Kcal / mol, but the main hydrophilic part in the b conformation is bound to the target small molecule target The b-conformation has low energy, and the main hydrophilic part in the b-conformation is bound to the hydrophilic end of the target small molecule target. Therefore, the b-conformation is desirable.
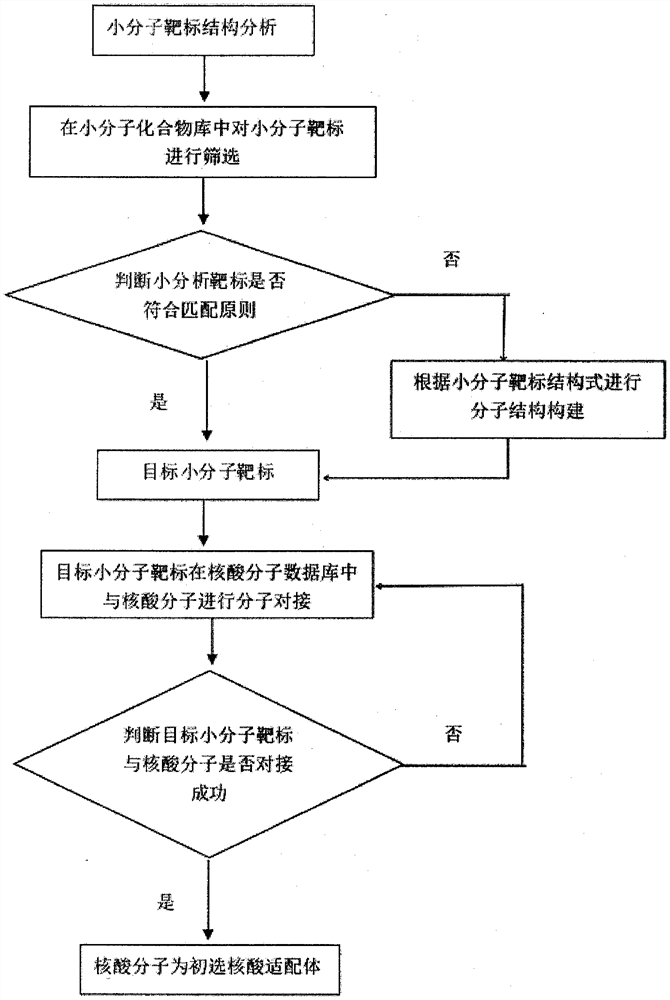
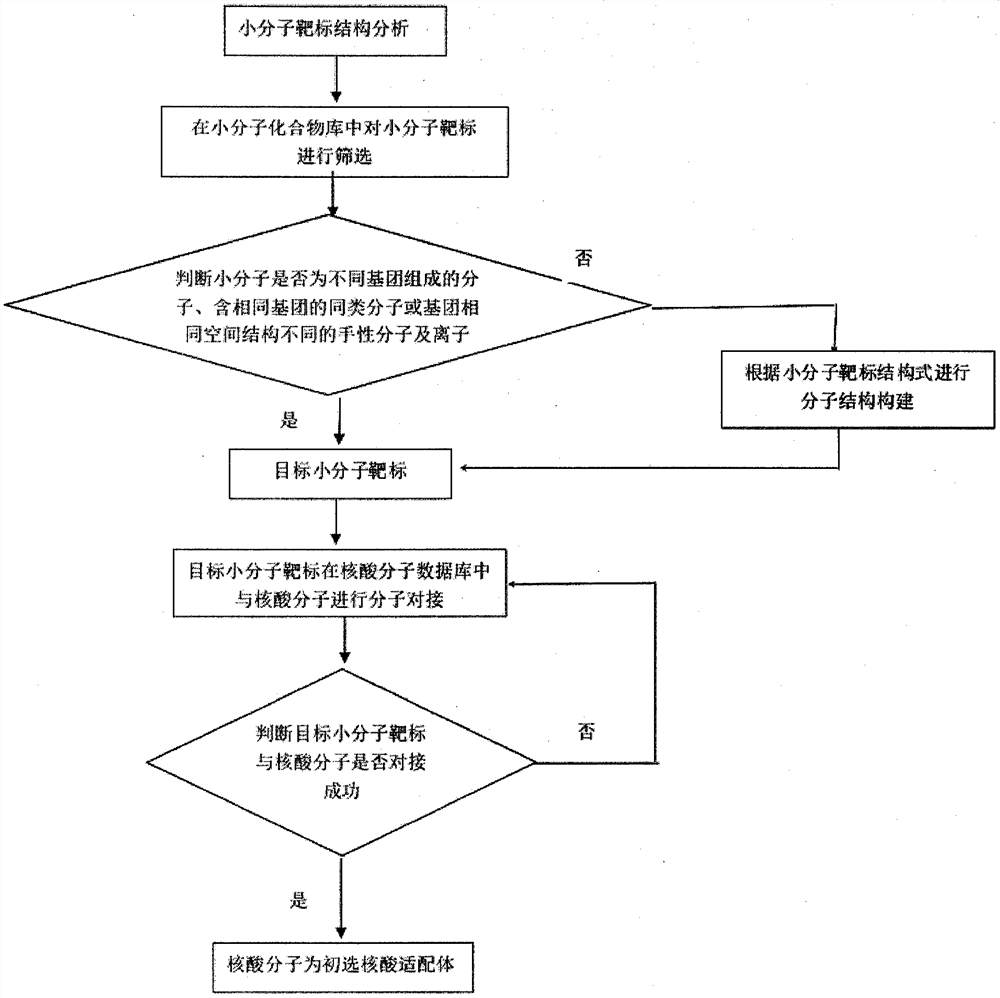
[0076] In a specific embodiment of the present invention, the search acceleration engine is used to screen the small molecule target in the small molecule compound library; the search acceleration engine is used to make molecular docking between the target small molecule target and the nucleic acid molecule in the nucleic acid molecule database. Construct an acceleration engine through parallelization technology to accelerate the matching and retrieval of eigenvalues of nucleic a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com