Yellow fluorescent protein and application thereof
A yellow fluorescent protein, fluorescent protein technology, applied in the biological field, can solve the problems of reporter gene application, incomplete, slow maturation of DsRed2, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0057] As a preferred embodiment, the promoter is CP25.
[0058] In the present invention, any random mutation method in the art can be used to introduce mutations, including (but not limited to) artificially synthesized degenerate primers to introduce random mutations; chemical shock-inducing agents to introduce random mutations; Random mutations were introduced into the strain by subculture; random mutations were introduced by mismatch PCR using the mismatch tendency of Taq polymerase. As a preferred embodiment, the method for introducing mutations is mismatch PCR or error-induced PCR, and the error-induced PCR amplification is performed by designing mismatched PCR primers.
[0059] As a more preferred embodiment, the error-causing PCR system includes: 1 μM upstream (downstream) primers, 0.2 mM dATP, 0.2 mM dGTP, 1 mM dCTP, 1 mM dTTP, 5.5 mM MgCl 2 , 0.1U / μl Taq DNA polymerase, 1ng / μl plasmid.
[0060] In the present invention, when constructing a recombinant plasmid, the ...
Embodiment 1
[0062] Construction and screening of embodiment 1 yellow fluorescent protein mutation
[0063] 1. Optimal expression of wild-type DsRed2 gene in E. coli
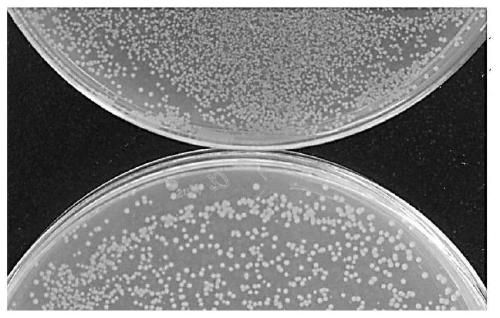
[0064] First, the DsRed2 (amino acid sequence shown in SEQ ID NO.1) gene was optimized according to the preferred codon of Escherichia coli, and the optimized gene was DsRed2E, which enabled the gene to be highly expressed in Escherichia coli, and the optimized amino acid sequence was the same as that of the wild type The sequence has not changed. Then add a CP25 promoter (the nucleic acid sequence is shown in SEQ ID NO.2) in front of the ORF of the gene to become a constitutive expression model, and the CP25 promoter activates the expression of the DsRed2 gene. Then cloned into the promoterless vector pBM16A vector. Screening was performed using chromogenicity of Escherichia coli. Colonies showing red are wild type.
[0065] 2. Error-prone PCR method to obtain mutant plasmids
[0066] The error-prone PCR method was use...
Embodiment 2
[0081] Example 2 Prokaryotic expression of DsYellow yellow fluorescent protein gene
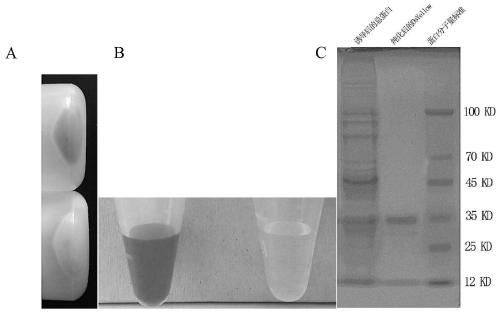
[0082] In order to further analyze the function of the obtained DsYellow yellow fluorescent protein gene, the DsYellow gene was constructed into the pBM30 vector by TOPO cloning, and the N-terminus of the DsYellow protein was tagged with 6His for purification. Induced expression was carried out by conventional methods, and purified with Ni-IDA magnetic agarose medium.
[0083] 1. Preparation and transformation of BL21(DE3) competent cells
[0084] Streak the BL21(DE3) strain frozen at -70°C on a non-resistant LB plate and culture it at 37°C overnight. The next day, pick a fresh BL21(DE3) single colony, inoculate it into 5 mL of LB medium, culture it with shaking at 37°C for 2-5 hours, and when the OD600 reaches about 0.5, take 1.5 mL of the bacterial liquid into a sterile centrifuge tube, Place in ice water bath for 10min. Centrifuge at 5000rpm for 2min at 4°C, discard the supernatant, and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com