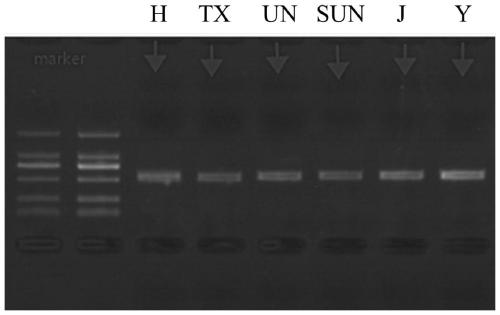
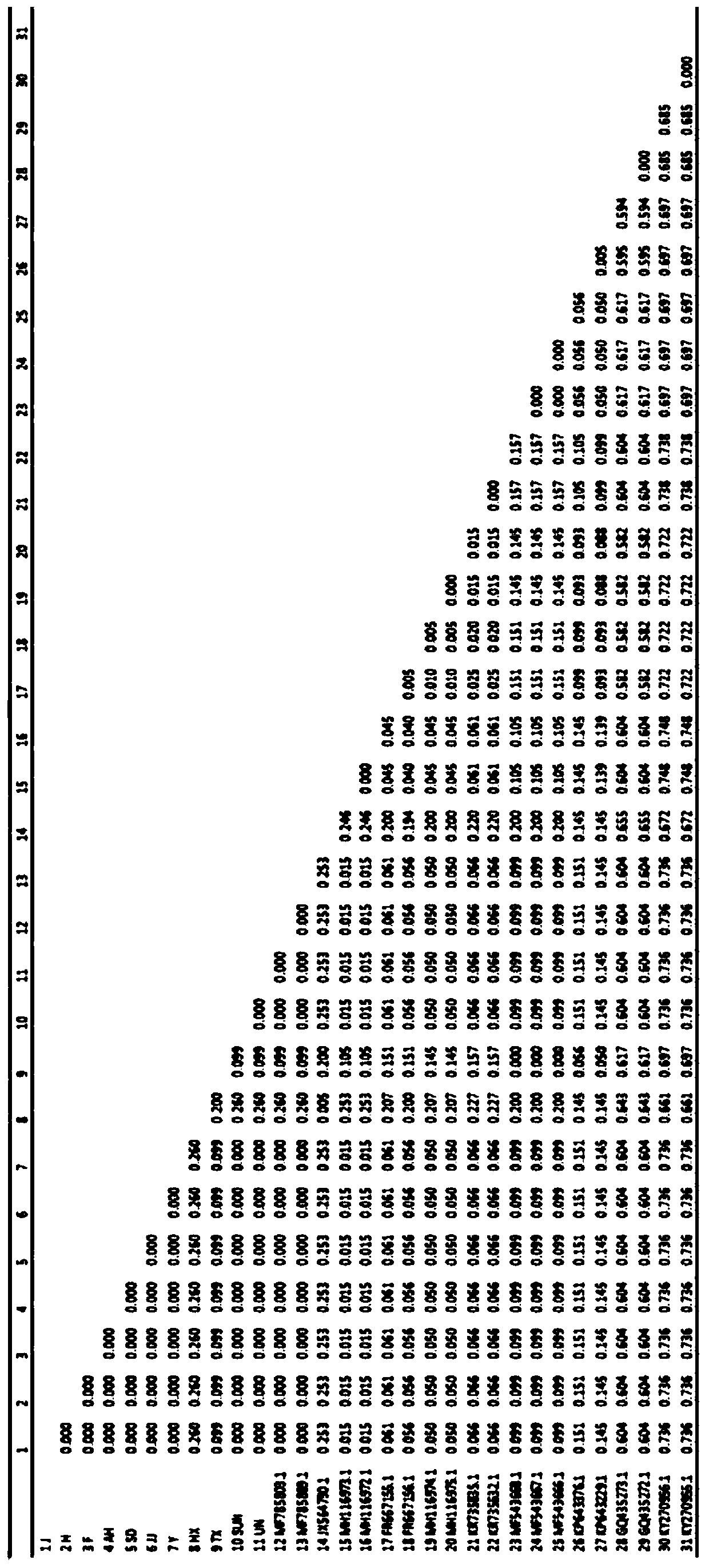
Method for identifying gnaphalium affine variety by utilizing psbA-trnH sequence
A technology for identifying Rat and Rhododendron, applied in the field of biological identification, can solve problems such as low accuracy, and achieve the effects of improving accuracy, good practical value and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] The present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments.
[0036] A kind of method of the present invention utilizes psbA-trnH sequence identification sagera species, specifically comprises the following steps:
[0037] Step 1, pre-treating the samples of S. japonicus to be identified, and extracting the DNA of the samples of S. japonicus to be identified using the Tiangen Plant Genomic DNA Extraction Kit, specifically:
[0038] Step 1.1, pretreatment of the unidentified sagebrush sample: take 30 g of the unidentified sagebrush sample that has been dried on silica gel, grind it into powder in a liquid nitrogen environment, and set aside;
[0039] Wherein the source of the samples to be identified is as shown in Table 1 below: including the sage in different regions, number 1-9, edelweiss, number 10, sagebrush, number 11,
[0040] Table 1 Sources of S. sageri samples to be identified
[0041]
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com