DNA data storage mixed error correction and data recovery method
An error correction and recovery method technology, applied in the data storage field of deoxyribonucleic acid, can solve the problems of high cost of digital information, waste of synthesis and sequencing resources, and reduced reliability of data recovery, so as to ensure reliability and improve the utilization of sequencing data. efficiency, high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045] In order to make the purpose, technical solution and advantages of the present invention clearer, the implementation manners of the present invention will be further described in detail below.
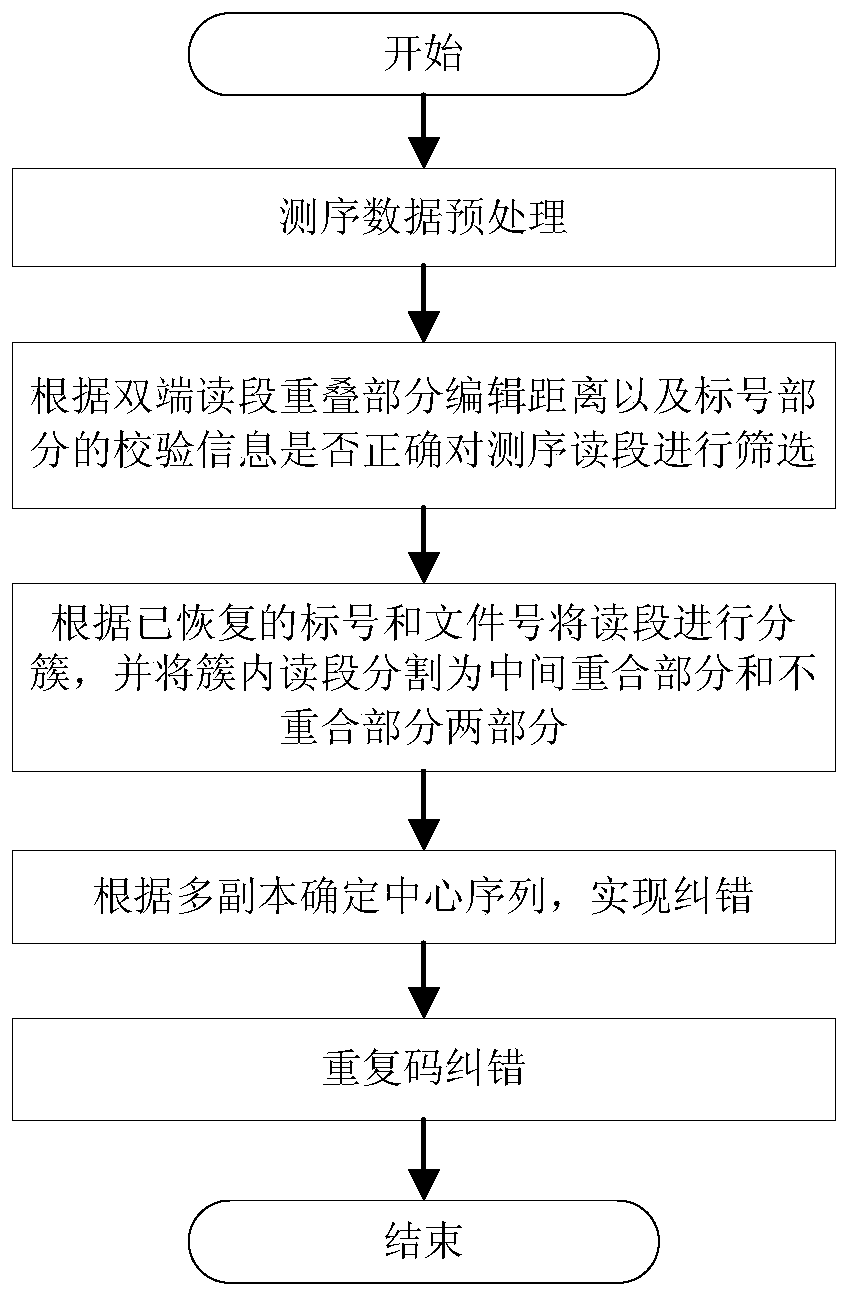
[0046] see figure 1 , a DNA data storage hybrid error correction and data recovery method, the method comprises the following steps:
[0047] (1) Preprocessing the sequencing data, specifically: for the double-end read sequence, one of the read segments is reversed and complemented to obtain two overlapping read segments;
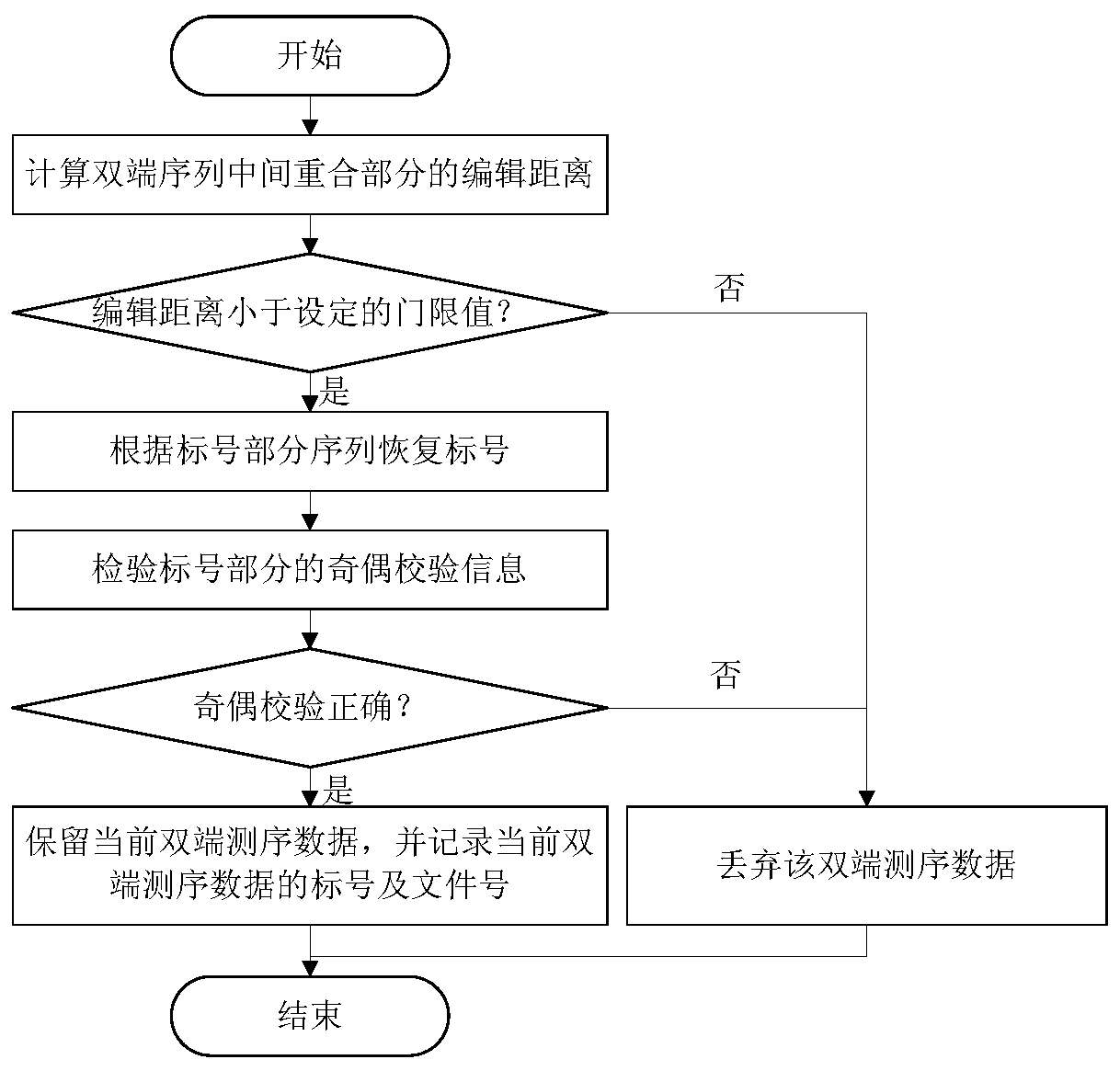
[0048] (2) Screen the sequencing reads according to the edit distance of the overlapping parts of the double-end reads and whether the verification information of the labeled part is correct;
[0049] (3) Cluster the reads according to the recovered label and file number, considering that the label part has been processed, and divide the reads in the cluster into two parts: the middle overlapping part and the non-overlapping part;
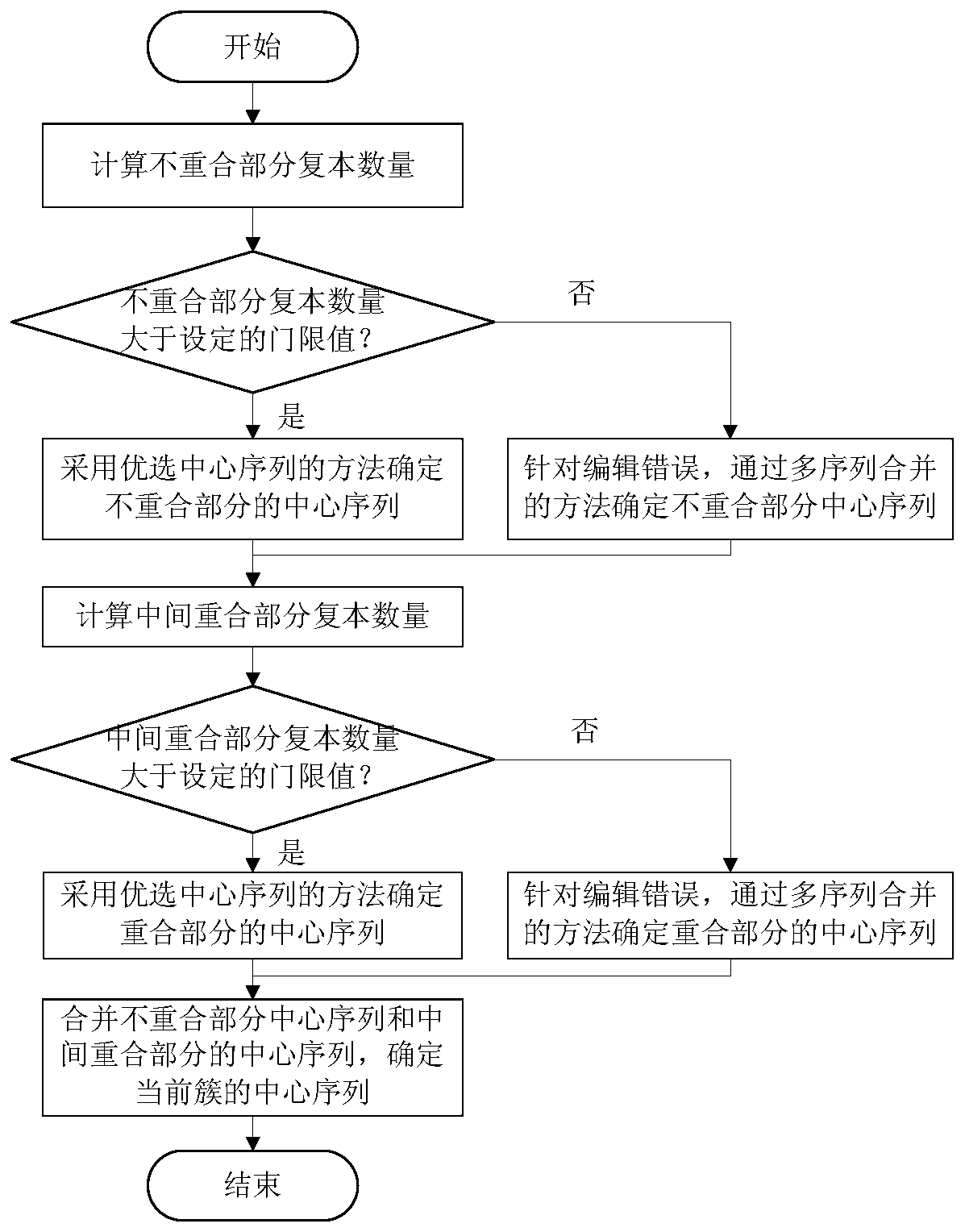
[0050] (4) If the copy n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com