Long non-coding RNA and application thereof
A long-chain non-coding, useful technology, applied in the field of biomedicine, can solve problems such as unclearness and changes in downstream cell functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Identification of long non-coding RNA and analysis of luciferase activity
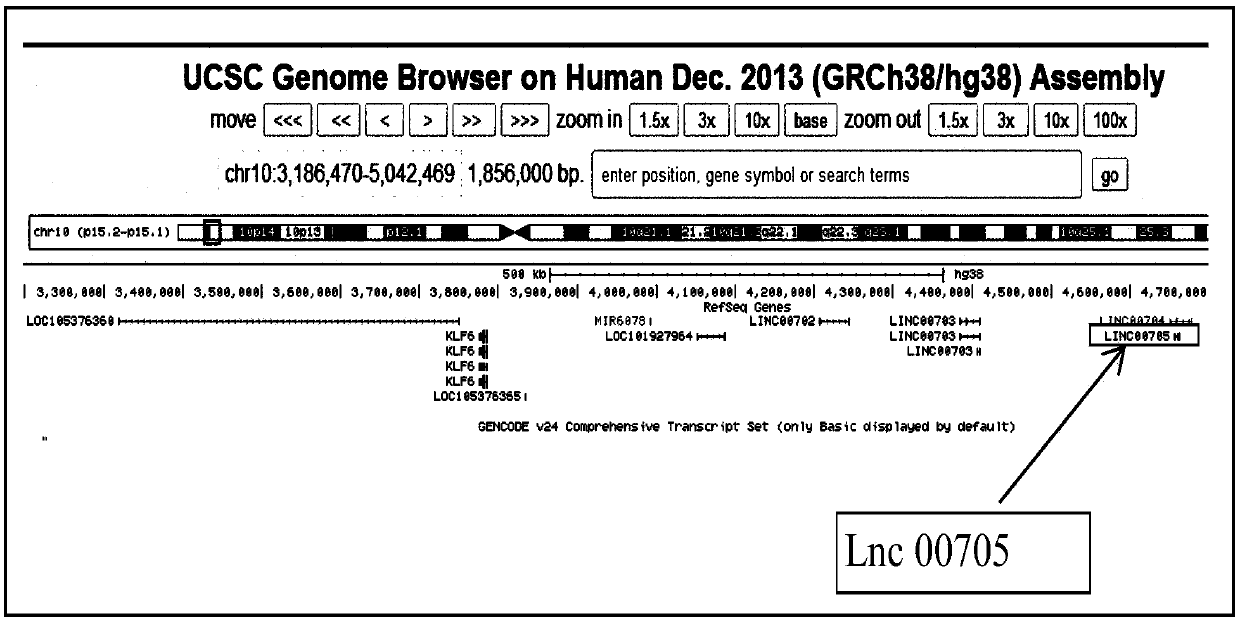
[0038] Using the UCSC online genome database analysis, it was found that there were a series of potential long-chain non-coding RNAs downstream of KLF6, as shown in Figure 1 (A), so there may be long-chain non-coding RNAs for feedback regulation of KLF6;
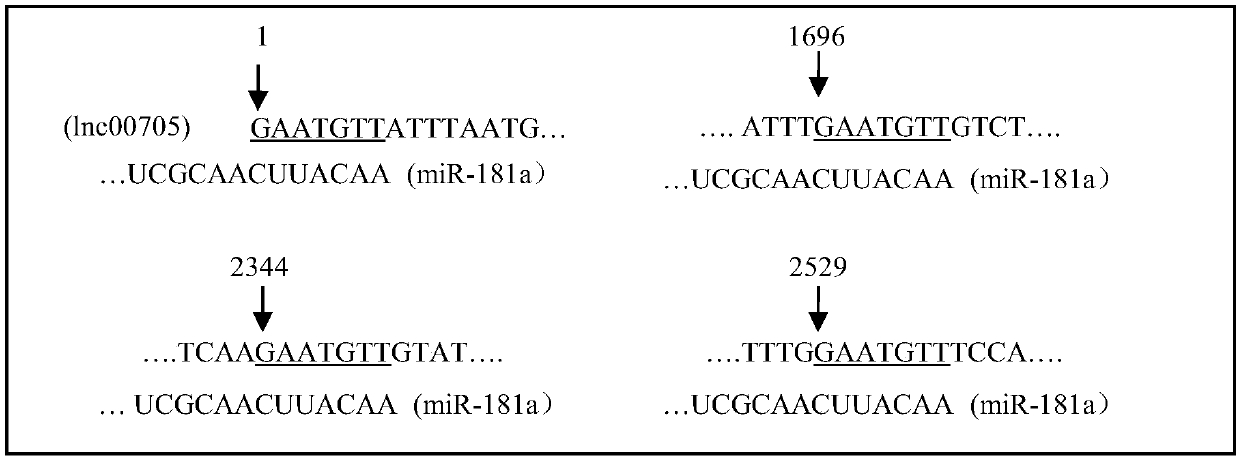
[0039] Based on previous reports, miR-181a has a targeted regulatory effect on KLF6; therefore, the analysis of long-chain non-coding RNA transcripts found that one of the long-chain non-coding RNA transcripts, Lnc00705, has four binding sites for miR-181a point, see Figure 1(B);
[0040] The nucleotide sequence of Lnc00705d is shown in SEQ ID No.1, and the nucleotide sequence of its transcript is shown in SEQ ID No.2.
[0041] SEQ ID No.1 is as follows:
[0042] GCGGAATATACGACTCACTATAGGGAGACCCAAGCTGGCTAGCGTTTAAACTTAAGCTTGCCACCTTTGATGTCTAGAATCAGGGGATCCGAATGTTATTTAATGGTGCCAATGATGTTGGAGGATACCTGTGGGAACCTGGATTAGAAGCATGTGCTGCTCTCAGACCTG...
Embodiment 2
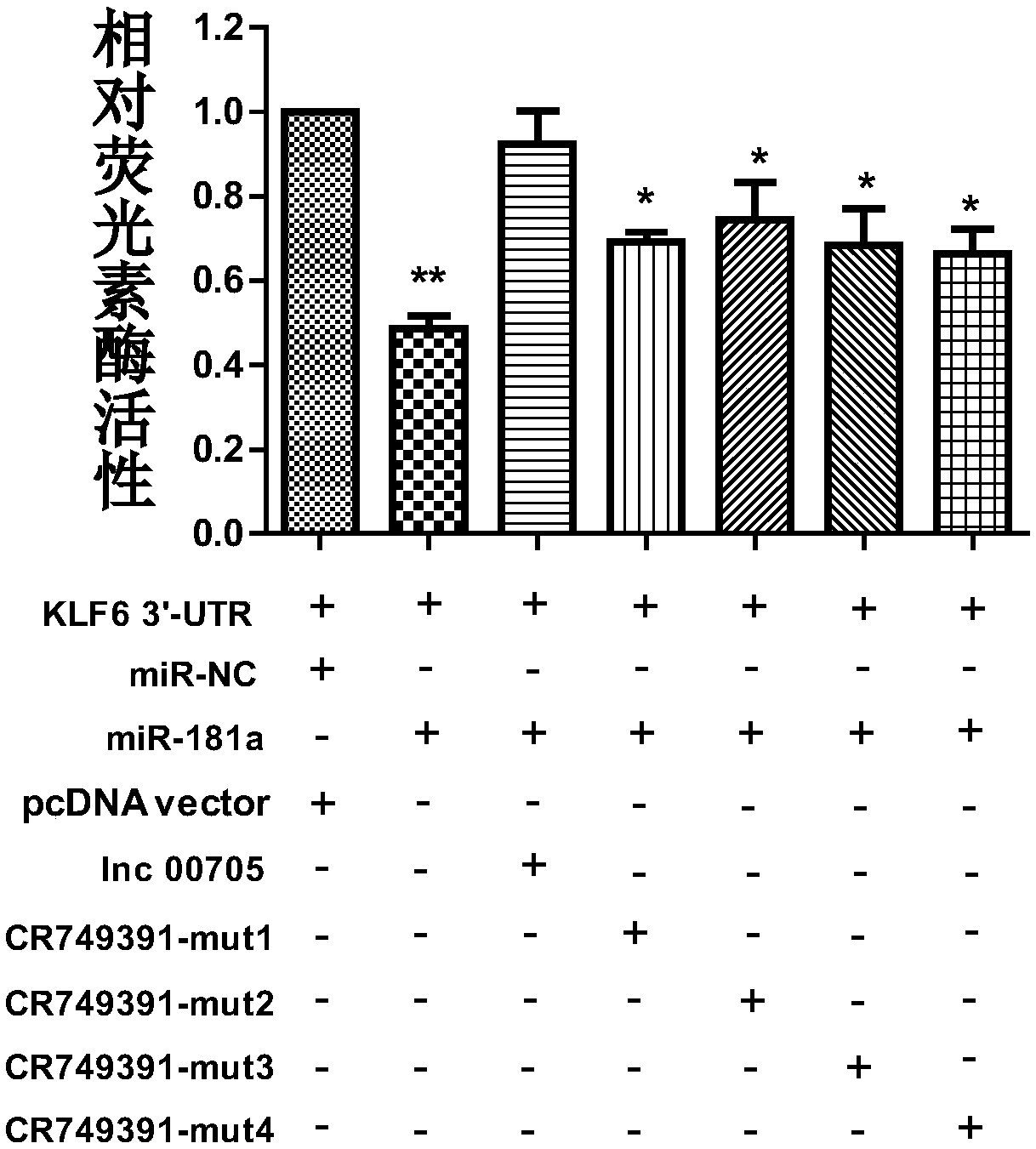
[0048] Example 2 Long non-coding RNA targets miR-181a to inhibit the expression of KLF6
[0049] The mucosal cell line GES-1 was used for routine culture. When the cell density reached 70%, Lnc00705siRNA was transfected with Lipofectamine 2000 (Invitrogen), and the medium was changed after 6 hours of transfection, and the culture was continued for 48 hours for the experiment;
[0050] The nucleotide sequence of Lnc00705siRNA is shown in SEQ ID No.3:
[0051] CUGACCAGCAUAGAUGUUA.
[0052] The detection of mRNA and protein expression levels was carried out by qRT-PCR and western blot respectively, and the results were as follows Figure 3(A)-Figure 3(C) shown;
[0053] Depend on Figure 3(A)-Figure 3(C) It can be seen that after inhibiting the expression of Lnc00705, miR-181a is upregulated, while the expression of KLF6 is down-regulated at both mRNA and protein levels.
Embodiment 3
[0054] Example 3 Effect of long non-coding RNA on proliferation and apoptosis of GES-1 cells
[0055] The effect of MTT method and flow cytometry on the proliferation and apoptosis of GES-1 cells after inhibiting Lnc00705, the results are as follows Figure 4(A)-Figure 4(C) shown;
[0056] Depend on Figure 4(A)-Figure 4(C) It can be seen that, compared with the control, inhibiting Lnc00705 promoted the proliferation of GES-1 cells and inhibited cell apoptosis.
[0057] In summary, the present invention provides a long-chain non-coding RNA and its application. The long-chain non-coding RNA is LncRNA00705, which is located downstream of the KLF6 gene. LncRNA00705 acts as a regulatory factor for diseases caused by miR-181a and KLF6 abnormalities, The medicine for preparing related diseases has broad application prospect and market value.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com