Method for monitoring reaction in synthetic DNA coding compound
A reaction monitoring and compound technology, applied in organic compound libraries, combinatorial chemistry, chemical libraries, etc., can solve the problems of different structures, different molecular weights, and difficulty in accurately monitoring the conversion degree of target compounds in mixed systems. Easy-to-use effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
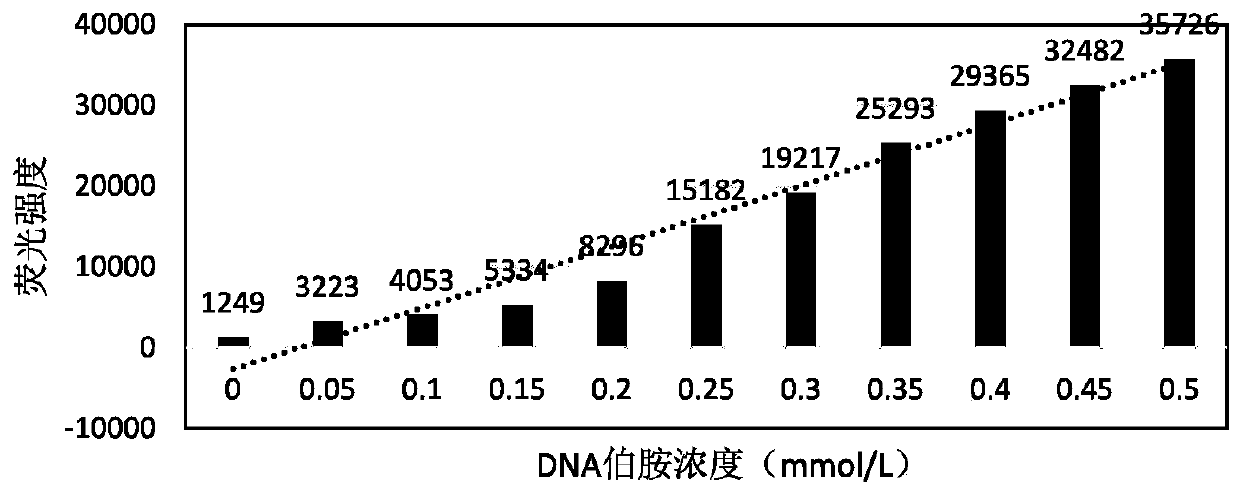
[0064] Embodiment 1, DNA-NH Concentration-absorption standard curve drawing of solution
[0065] Take DNA-NH2 solution with an initial concentration of 0.5mmol / L, and prepare standard solutions with concentrations of 0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, 0.40, 0.45, 0.50mmol / L. Add the acetone solution (50mmol / L) of the fluorescent reagent 4-phenylspiro[furan-2(3H), 1-dihydroisobenzofuran]-3,3′-dione (50mmol / L) in the amount of 10 times the substance, 5°C Incubate for 10 minutes. Detect fluorescence absorption at an excitation wavelength of 388nm and an emission wavelength of 475nm, and draw an absorption-concentration standard curve, such as figure 1 shown.
Embodiment 2
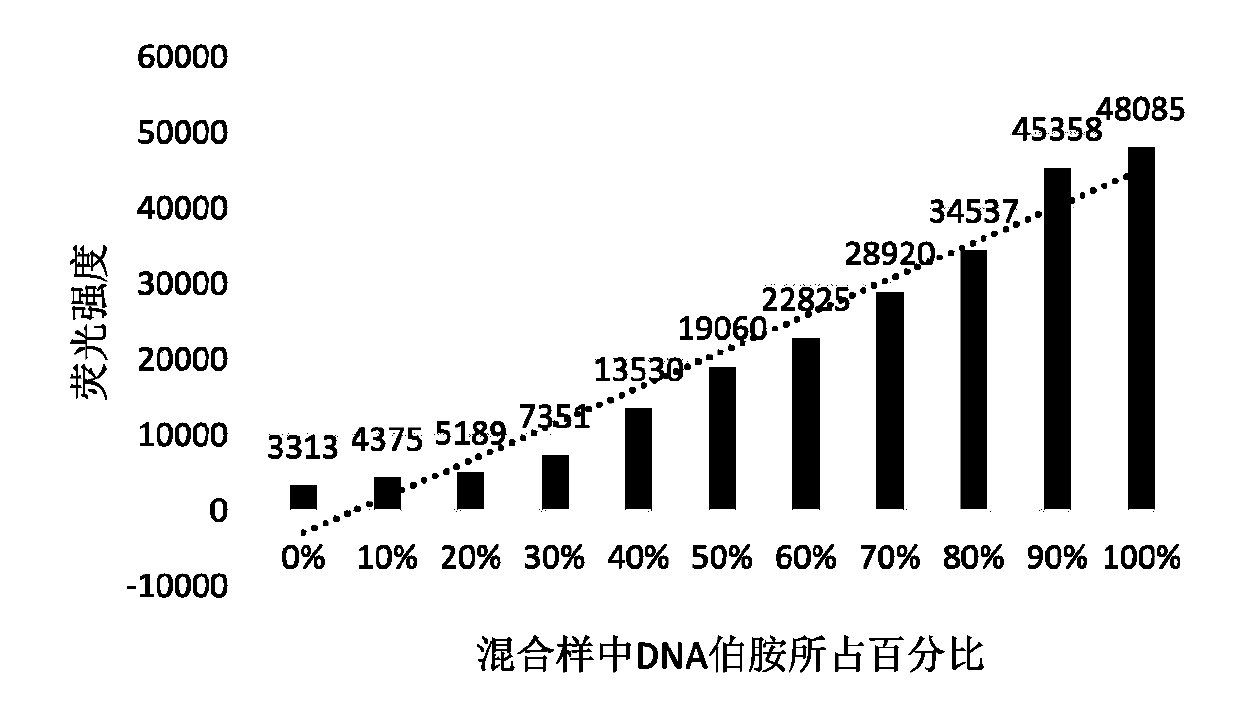
[0066] Example 2, the drawing of the concentration-absorption standard curve of DNA-NH2 solution in the mixed system
[0067] Get the DNA-NH2 solution and DNA-NH solution that initial concentration is 0.5mmol / L, mix and prepare DNA-NH2 ratio (mol) be 10%, 20%, 30%, 40%, 50%, 60%, 70% , 80%, 90% solution. Add 50 times (substance amount) fluorescent reagent acetone solution (50mmol / L), and incubate at 40°C for 5 minutes. Detect fluorescence absorption at an excitation wavelength of 388nm and an emission wavelength of 475nm, and draw an absorption-concentration standard curve, such as figure 2 shown.
Embodiment 3
[0068] Example 3. Monitoring of the reaction process in the complex system of the DNA-encoded compound library
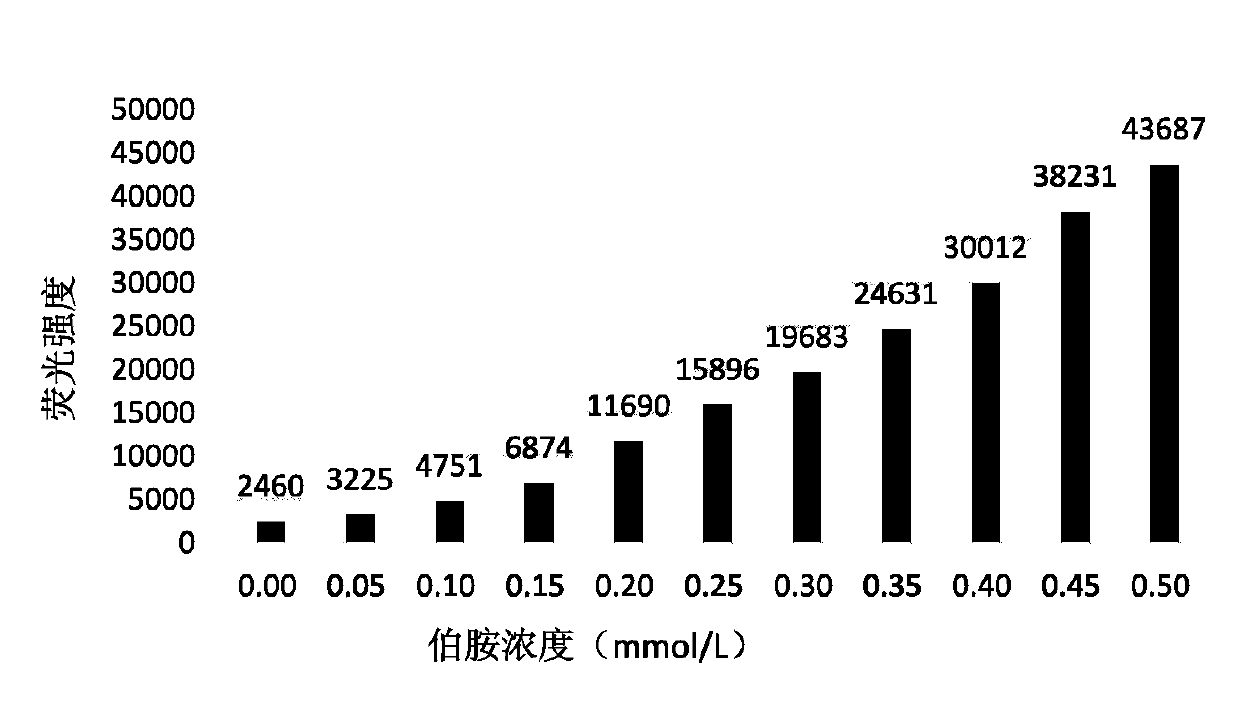
[0069] 1. Construct a standard curve using a single DNA with a similar backbone
[0070] Get the 1b (structural formula such as Figure 5 Shown) solution, prepared as a standard solution with a concentration of 0.05, 0.10, 0.15, 0.20, 0.25, 0.30, 0.35, 0.40, 0.45, 0.50mmol / L. Add 20 times (substance amount) fluorescent reagent acetone solution (50mmol / L), and incubate at room temperature for 10 minutes. Fluorescent absorption was detected at an excitation wavelength of 388nm and an emission wavelength of 475nm, and an absorption-concentration standard curve was drawn. Such as image 3 shown
[0071] 2. Single DNA reaction monitoring process (such as Figure 4 )
[0072] 2.1 1a to 1b reactions (eg Figure 5 )
[0073] Dissolve 1a in sodium borate buffer (pH=9.4, concentration 250mmol / L) to prepare a 1mmol / L solution, and dissolve 1a with tris(2-carboxyethyl)p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com