Primer and probe set for detecting helicobacter pylori drug-resistant mutation sites and application thereof
A drug-resistant mutation site, Helicobacter pylori technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/testing, etc., can solve the problems of low sensitivity, time-consuming, complicated operation, etc., and achieve high sensitivity , short time-consuming effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Embodiment 1, primer, probe design
[0051] By analyzing the sequences of Helicobacter pylori amoxicillin resistance gene PBP1A and tetracycline resistance gene 16SrRNA, drug resistance mutation sites were selected, and specific primer probes were designed, as shown in Table 1 below.
[0052] Table 1
[0053] SEQ ID NO.1 PBP-F1 aacaaaaccacgcatggcaccccaca SEQ ID NO.2 PBP-F2 aaaaccgggacttctaacaaat SEQ ID NO.3 PBP-F3 tttagaaatcgccggtaaatg SEQ ID NO.4 PBP-block1 agtgaacaaaaccacgcatggcaccccagc SEQ ID NO.5 PBP-block2 tcgccggtaaaaccgggacttctaacaaca SEQ ID NO.6 PBP-block3 cattaaaggtttagaaatcgccggtaaaac SEQ ID NO.7 PBP-R gcgttcctcgctatcgtctg SEQ ID NO.8 PBP-P acgatttctttacgcaagcctttggggacatc SEQ ID NO.9 16S-F agcatgtggtttaattcgattc SEQ ID NO.11 16S-block agcggtggagcatgtggtttaattcgaaga SEQ ID NO.10 16S-R cgctcgttgcggggactta SEQ ID NO.12 16S-P ccaacatctcacgacacgagctgacga...
Embodiment 2
[0058] Embodiment 2, fluorescent PCR
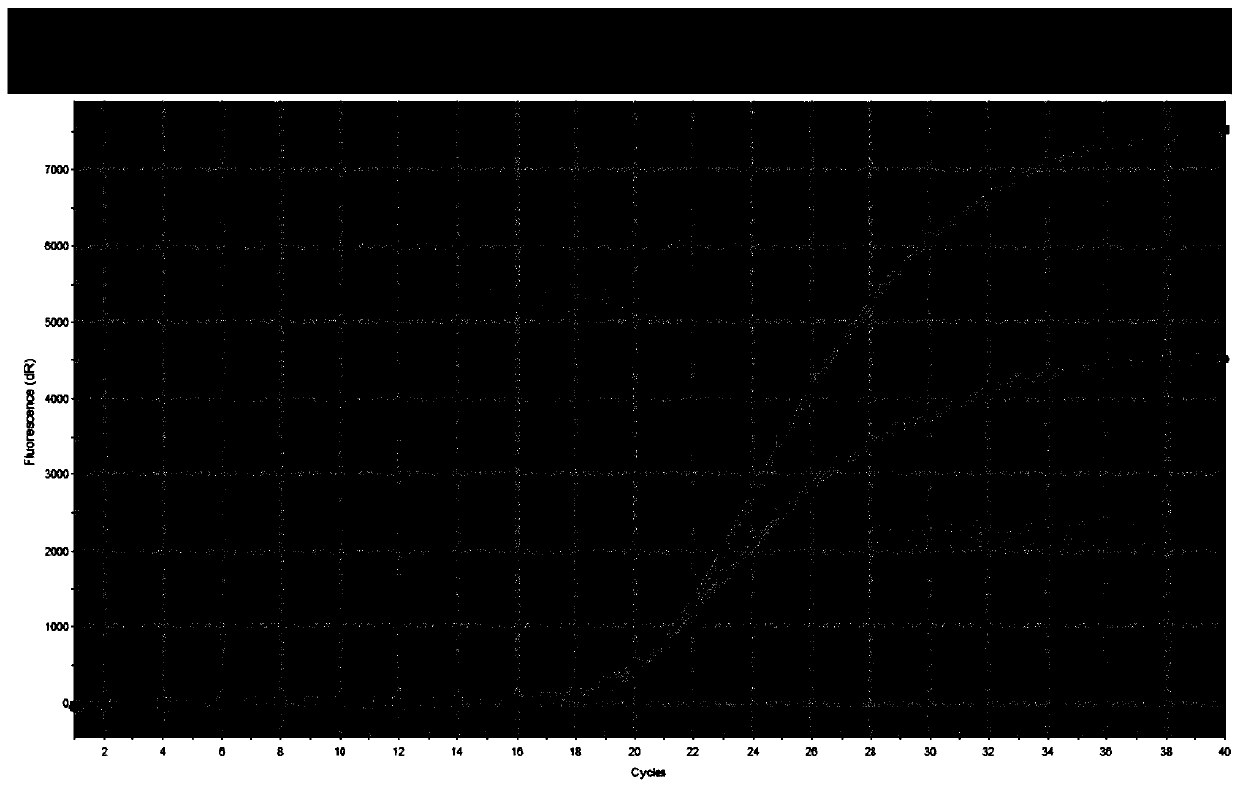
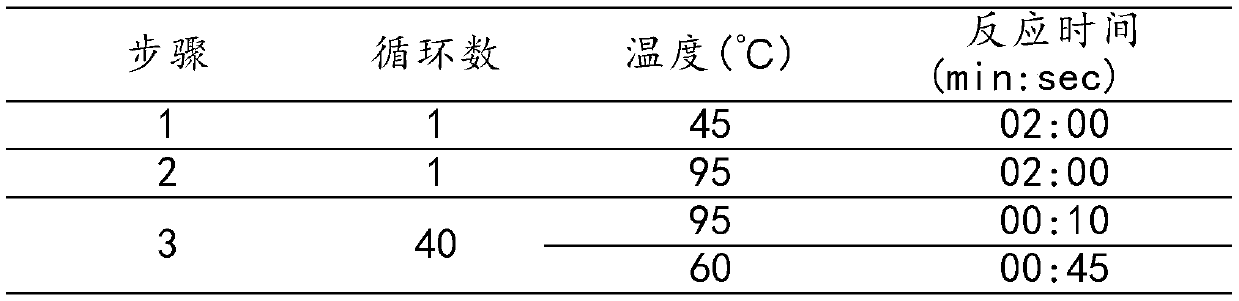
[0059] The primers, blocking sequences and probes provided in Example 1 can be used to perform fluorescent PCR to detect drug resistance mutation sites of Helicobacter pylori.
[0060] Table 2 shows the formula of PCR reaction solution.
[0061] Table 2
[0062] Reagent name Volume (μL) 10×PCR Buffer (Mg 2+ Plus)
2.5 MgCl 2 (1M)
0.05 dNTPs [contains dUTP] (25mM) 0.2 PBP-F1 0.6 PBP-F2 0.6 PBP-F3 0.6 PBP-block1 1.2 PBP-block2 1.2 PBP-block3 1.2 PBP-R 0.6 PBP-P 0.3 16S-F 0.6 16S-block 1.2 16S-R 0.6 16S-P 0.3 HS Taq (5U) 0.4 UNG enzyme (1U) 0.1 Sterile RNase-free water 7.75 template 5 total 25
[0063] Wherein, in the prepared reaction solution, the concentration of each blocking sequence is twice the concentration of the respective corresponding primers. The optimal concentration of each primer was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com