Method for rapidly predicting heterosis based on genome size of crops
A technology of heterosis and genome, applied in the direction of plant genetic improvement, botanical equipment and methods, applications, etc., can solve the problems of inability to reproduce through grafting, achieve shortening the breeding cycle, strong operability, and reduce breeding costs Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
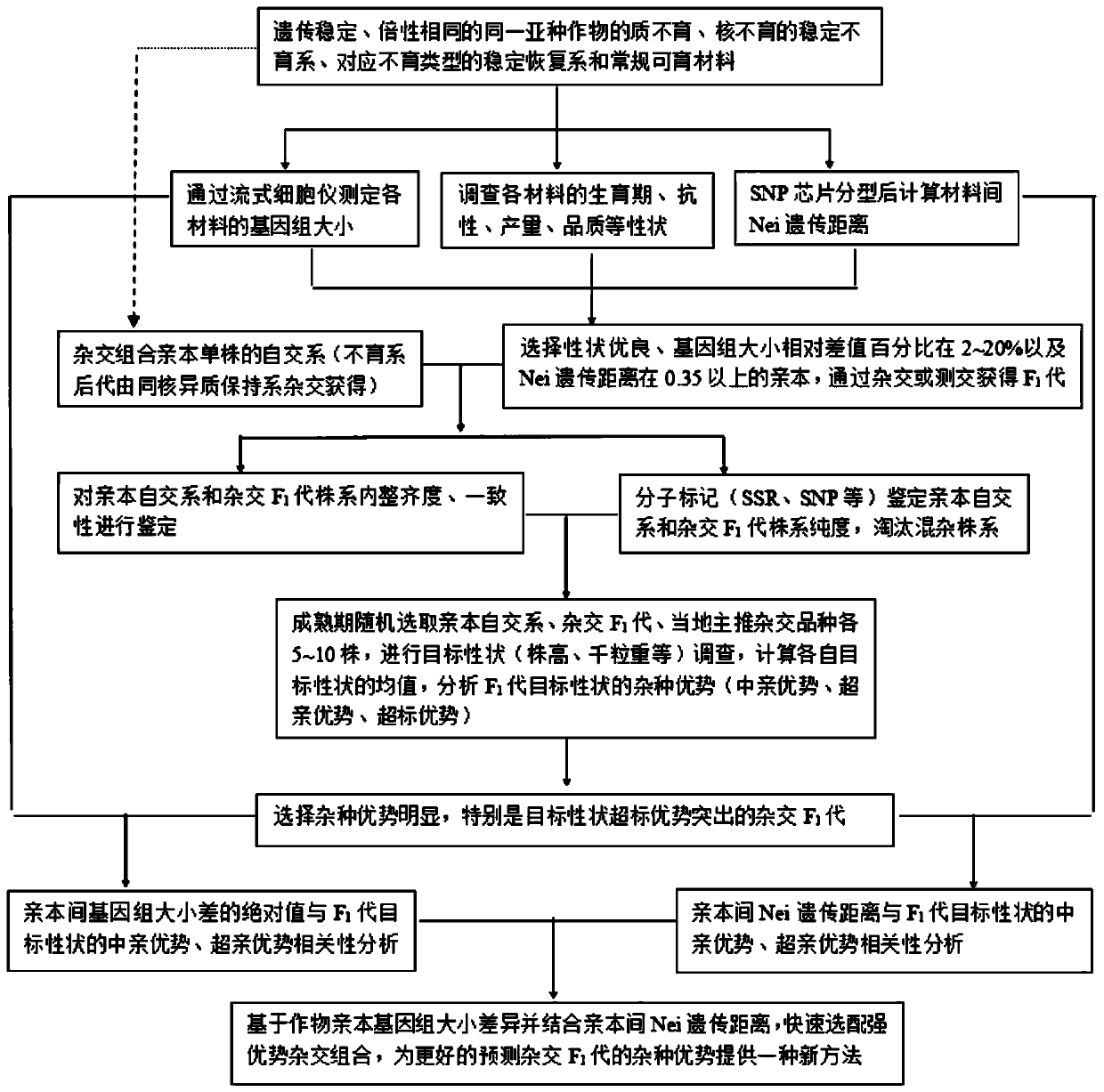
[0049] Such as figure 2 As shown, taking Brassica napus as an example, this embodiment provides a method for quickly predicting heterosis based on genome size, including the following steps:
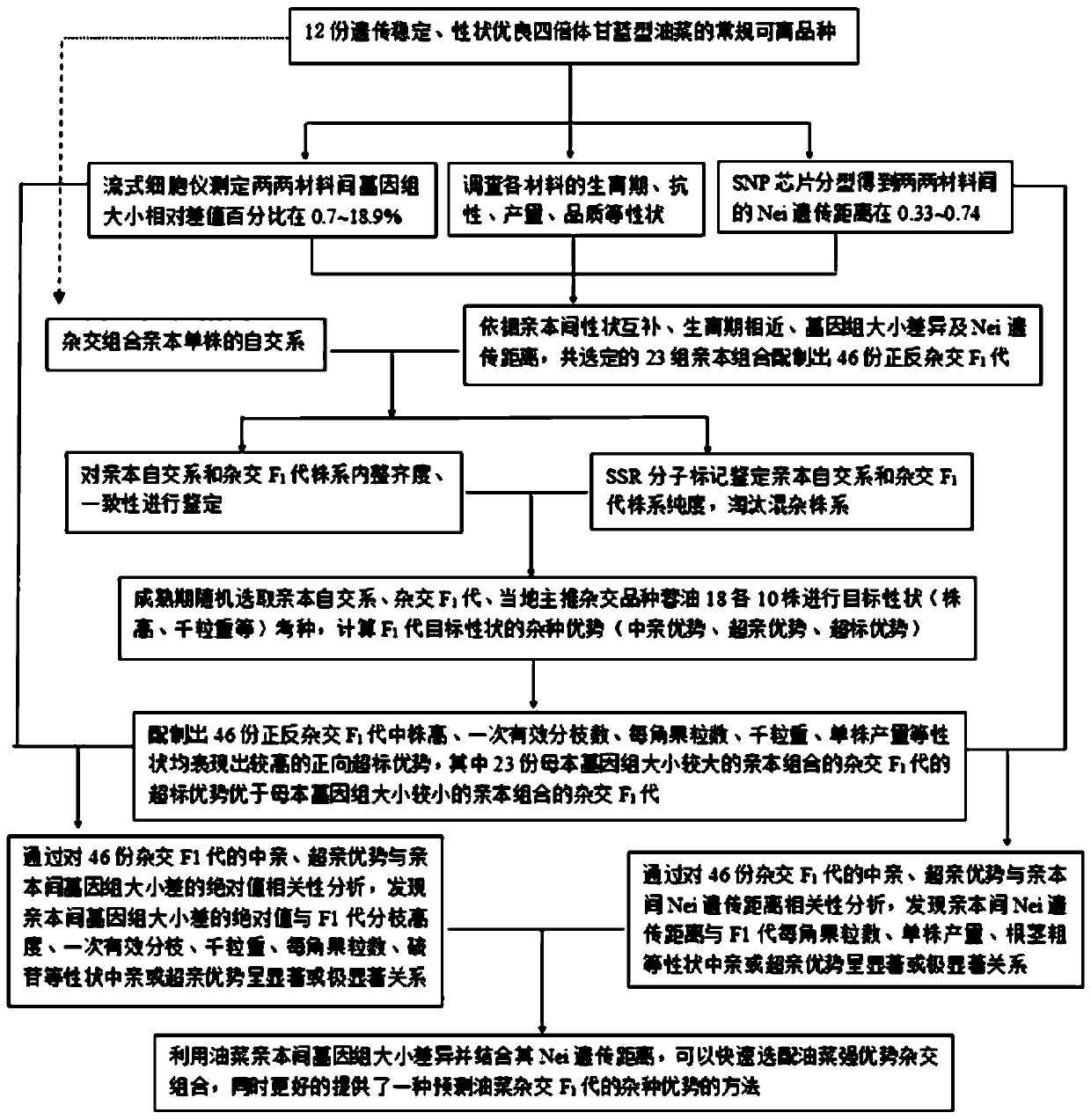
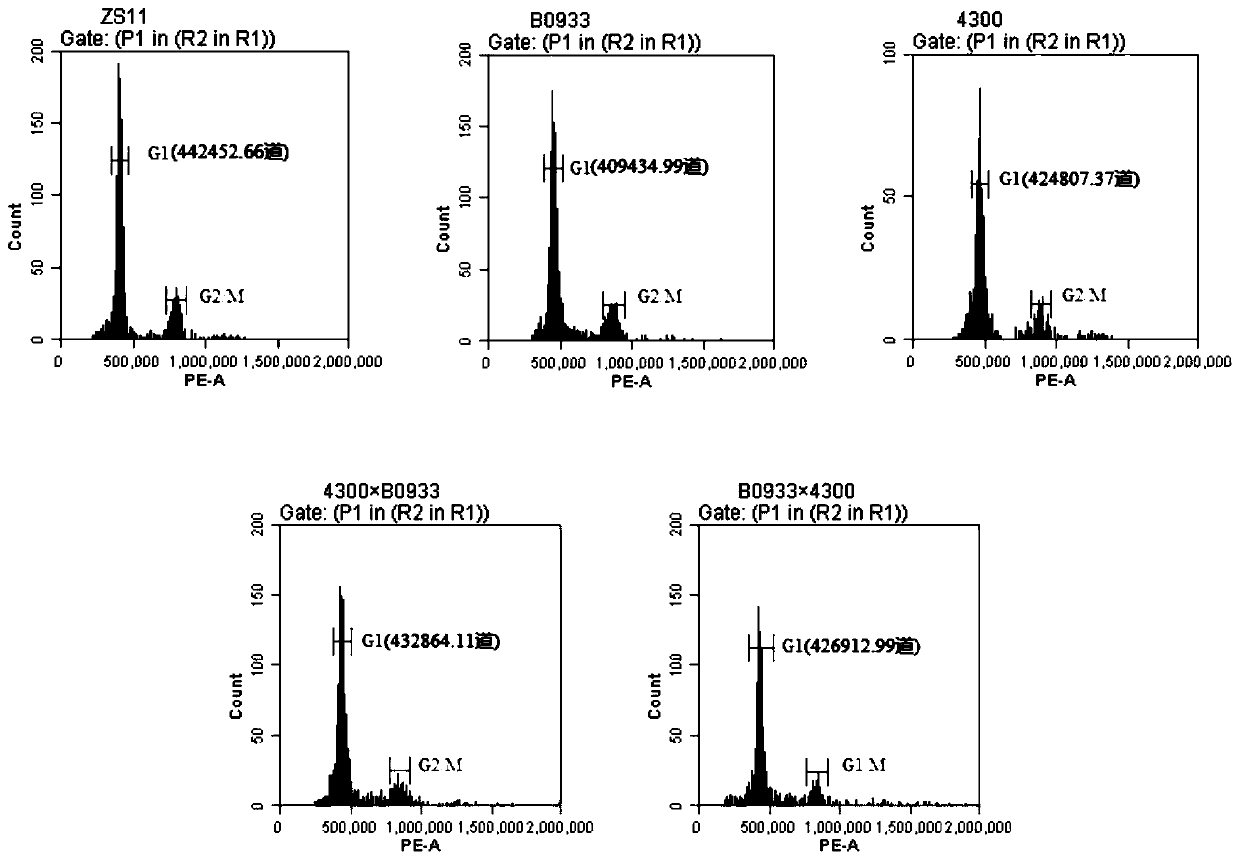
[0050] S1. Using 12 genetically stable tetraploid Brassica napus as parent crops, measure the genome size of these 12 materials by flow cytometry, and calculate the relative difference percentage of genome size between any pair of materials in the 12 materials In the range of 0.7 to 18.9%. The Nei genetic distance between pairs of materials was found to be in the range of 0.33-0.74 by SNP chip typing. Among them, there are 48 hybrid combinations that meet the conditions of a. The relative difference percentage of the genome size of the two parent crops is 2-20% and b. The Nei genetic distance of the two parent crops is greater than or equal to 0.35, and then combine 12 materials traits (growth period, resistance, yield, quality and other traits), a total of 23 groups of hybrid combina...
Embodiment 2
[0059] Such as Figure 5 As shown, taking rice as an example, this embodiment provides a method for rapidly predicting heterosis based on genome size, including the following steps:
[0060] S1. 16 genetically stable diploid indica varieties were used as parent crops. The genome size of these 16 materials was determined by flow cytometry. The relative difference percentage of genome size between pairs of materials in 16 materials was calculated in the range of 0.3-15.2%, and the Nei genetic distance between pairs of materials was in the range of 0.24-0.67 through rice 50K SNP microarray typing. Among them, there are 53 hybrid combinations that meet the conditions a. The relative difference percentage of the genome size of the two parent crops is 2-20% and b. The Nei genetic distance of the two parent crops is greater than or equal to 0.35, and then 16 materials are combined. Traits (growth period, resistance, yield, quality and other traits), screen out consistent growth per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com