High-throughput prediction method for neoantigens of pan-cancer tumor and application thereof
A prediction method and tumor technology, applied in the fields of bioinformatics and tumor immunotherapy, can solve problems such as time-consuming and labor costs, and difficult problems of tumor neoantigen screening methods, so as to reduce workload, save prediction time, and reduce redundancy The effect of steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1 Prediction of tumor neoantigens
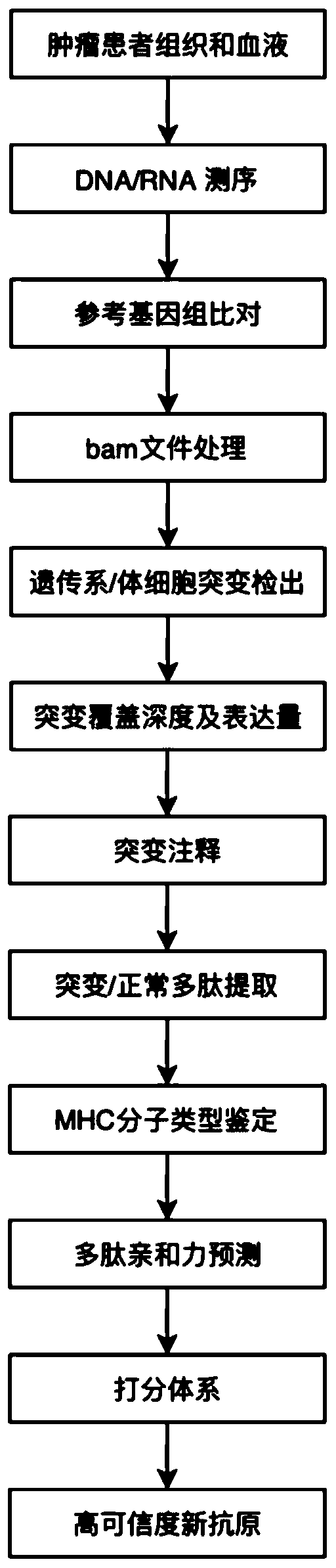
[0058] The flow chart of the present invention predicting tumor neoantigens is shown in figure 1 shown. The detailed process is as follows:
[0059] 1. Material preparation
[0060] The tumor tissue of the patient with tumor number AO001 (patient with hepatocellular carcinoma) was obtained, and WES and RNA-seq sequencing of the tumor tissue were completed through the illumina high-throughput sequencing platform.
[0061] 2. Data quality control
[0062] The original fastq data of DNA and RNA sequencing were quality controlled by FastQC software to obtain the data AO001.clean.fq.gz after quality control filtering.
[0063] 3. Data comparison
[0064] The DNA data after quality control was compared with the reference genome using BWA software to obtain bam files of tumor and normal tissue DNA data, and the RNA after quality control was compared with the reference genome using hisat2 software to obtain bam files of tumor RNA...
Embodiment 2
[0087] Example 2 Candidate tumor neoantigen verification
[0088] According to the scoring order in Table 1 in Example 1, some tumor neoantigens were selected to undergo tetramer verification experiments to test the accuracy and reliability of the prediction method of the present invention.
[0089] Steps: experiment according to QuickSwitch TM The operating instructions of the quant tetramer kit were carried out.
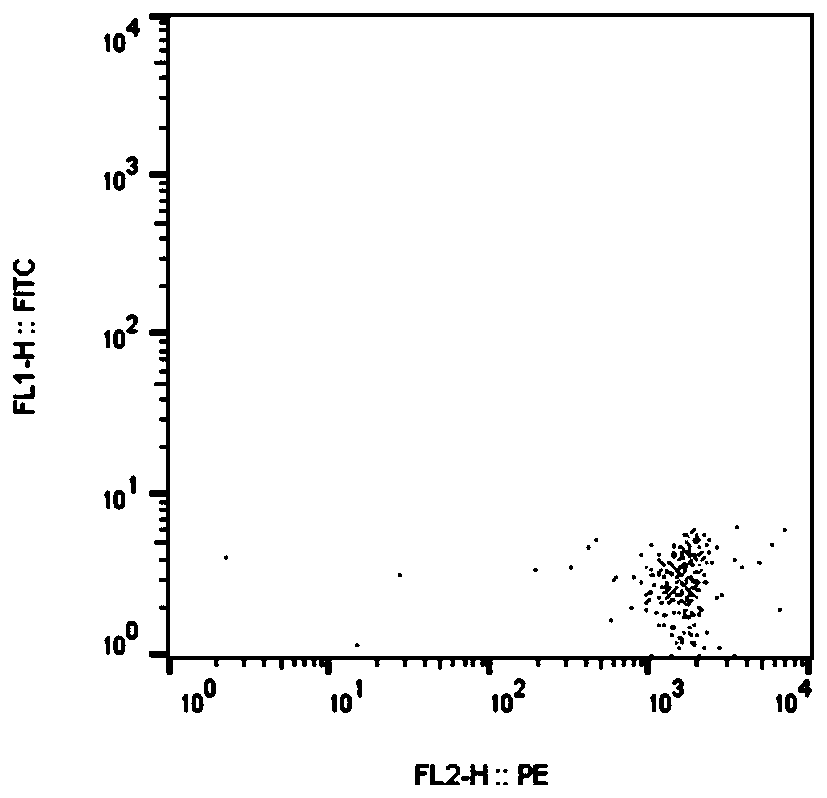
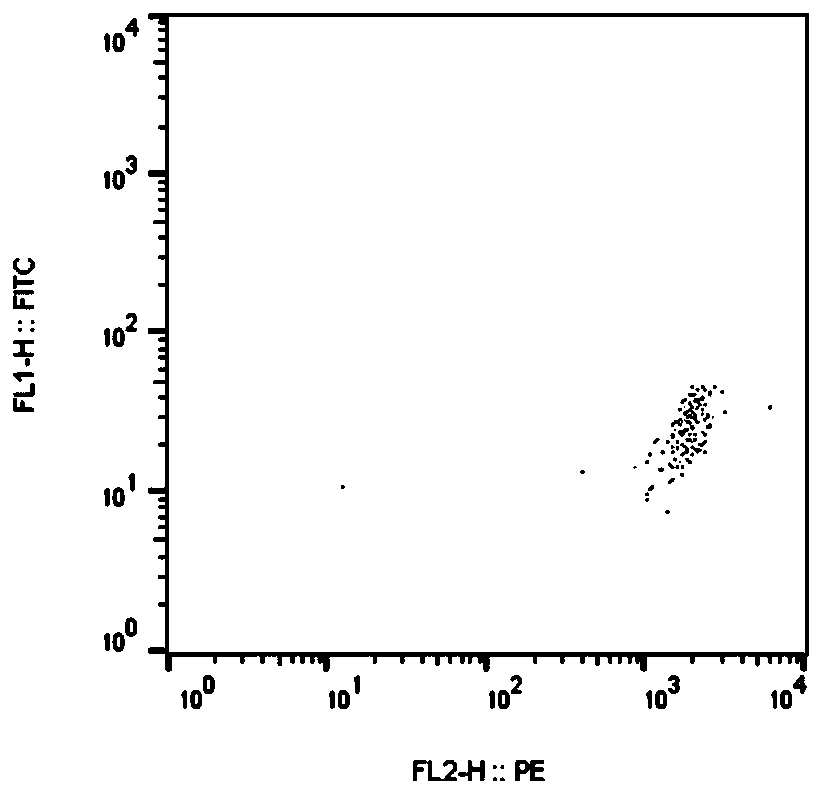
[0090] Results: 5 positive peptides were obtained, and the 5 positive peptides were: SLK, ETAA1, DOCK7, CYP2C8, TPR, Figure 4-8 It represents the results of flow cytometry detection of the above five positive peptide tetramer displacement experiments. Figure 2-3 Respectively are positive control polypeptide, negative control polypeptide tetramer displacement experiment flow cytometer detection results.
[0091] It can be seen that the positive polypeptides obtained in the verification are tumor neoantigens with high scores evaluated by the prediction method of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com