Escherichia coli engineering bacterium for producing 2'-fucosyllactose
A technology of fucosyllactose and Escherichia coli, which is applied in the direction of glycosyltransferase, bacteria, biochemical equipment and methods, etc., can solve the problem of high price of L-fucose and low yield of 2'-fucosyllactose Low, difficult fermentation production and other problems, to achieve the effect of increasing yield, increasing yield, and relieving metabolic pressure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
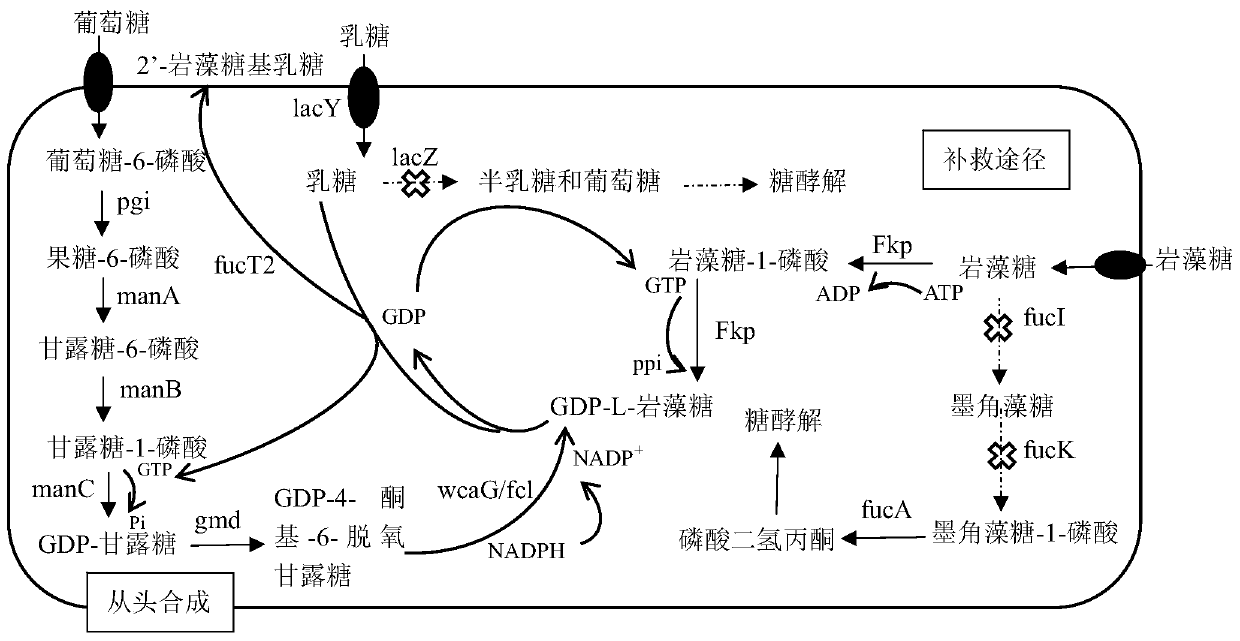
[0030] Example 1 Escherichia coli BL21 wacJ, lacZ, fucIK gene knockout
[0031] Using the CRISPR-Cas9 gene knockout system to knock out wacJ, lacZ, and fucIK in Escherichia coli BL21, the specific steps are as follows (see Table 1 for the primer sequences involved):
[0032] (1) Using the Escherichia coli BL21 genome as a template, using wcaJ-up-F / R and wcaJ-down-F / R, lacZ-up-F / R and lacZ-down-F / R, fucIK-up-F / R and fucIK-down-F / R were amplified by PCR to amplify the upstream and downstream fragments of wacJ, lacZ, and fucIK, respectively, and the gel was recovered. Then use wacJ, lacZ, fucIK upstream and downstream fragments as templates respectively, using wcaJ-up-F / wcaJ-down-R, lacZ-up-F / lacZ-down-R and fucIK-up-F / fucIK-down-R The primers were used to obtain complete wacJ, lacZ, and fucIK templates by inversePCR, and the DNA fragments were recovered from the gel.
[0033](2) Using the original pTargetF plasmid as a template, wcaJ-sg-F / R, lacZ-sg-F / R and fucIK-sg-F / R as pr...
Embodiment 2
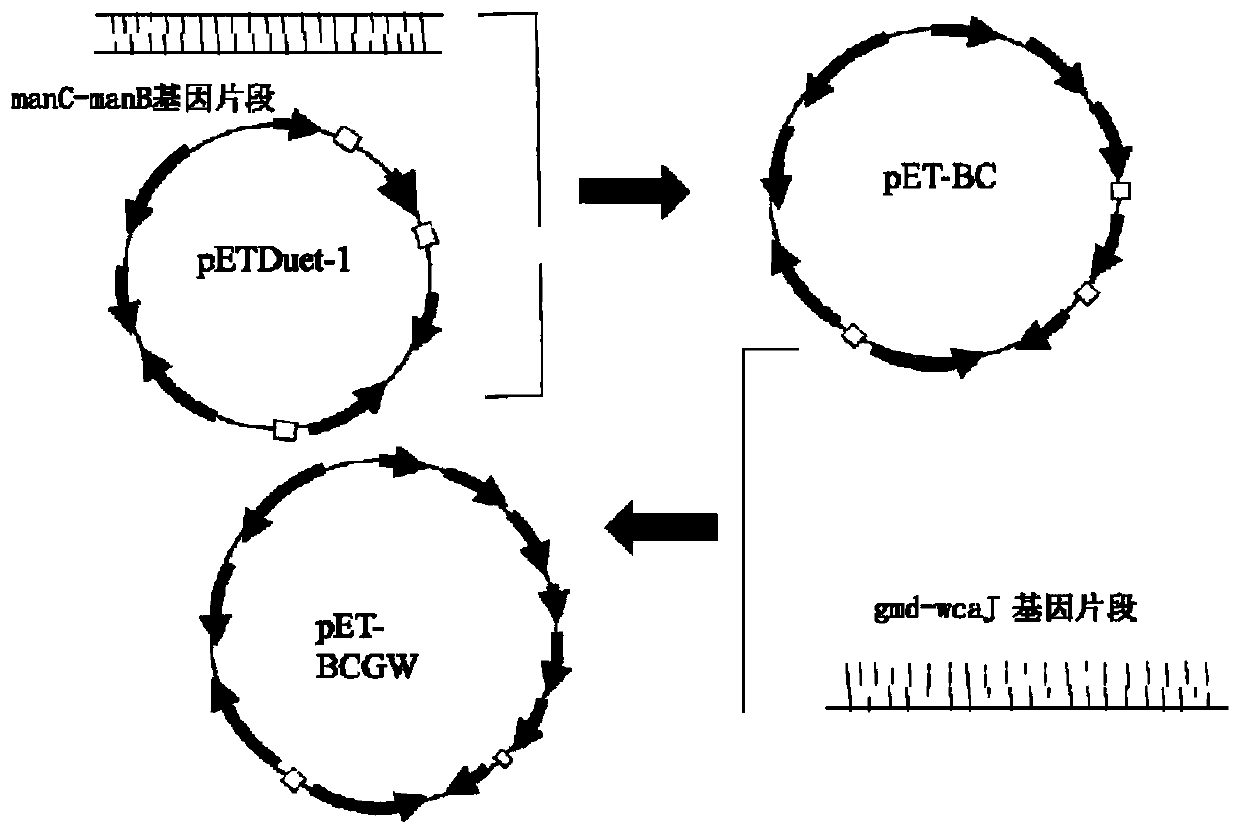
[0041] The construction of embodiment 2 recombinant expression vector
[0042] The specific steps for constructing the recombinant expression vector are as follows (see Table 2 for the primer sequences involved):
[0043] (1) Obtaining of manC-manB and gmd-wacG gene cluster fragments: using the genome of Escherichia coli K-12 (Escherichiacoli) as a template, using manCB-F / R (NcoI) and GW-F / R (NdeI) as primers , the manC-manB and gmd-wacG gene cluster fragments were amplified by PCR, and the DNA fragments were recovered from the gel;
[0044] (2) Obtaining the fkp gene fragment: using the Bacteroides fragilis 9343 genome as a template and using Fkp-F / R (NdeI) as a primer, the fkp gene fragment was amplified by PCR, and the DNA fragment was recovered from the gel
[0045] (4) Obtaining the fucT2 gene fragment: using Helicobacter pylori (Helicobacter pylori) genome as a template, using FucT2-F / R (NcoI) as a primer, amplify the fucT2 gene fragment by PCR, and recover the DNA frag...
Embodiment 3
[0050] The construction of embodiment 3 escherichia coli engineering strain
[0051] Cultivate wacJ, lacZ, fucIK gene knockout strain BWLF and prepare competent cells, use the chemical transformation method to introduce the extracted plasmids pET-BCGW and pCD-FF into the strain, and put them on the double-antibody LB plate (ampicillin and streptomycin) 37 Cultivate overnight at ℃ to obtain genetically engineered bacteria producing 2'-fucosyllactose. The construction of other recombinant genetically engineered bacteria is as above, and the specific recombinant plasmids, engineered bacteria and their detailed information are shown in Table 3.
[0052] Table 3 Details of plasmids and engineered bacteria
[0053]
[0054]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com