DNA methylation biomarker composition, detection method and kit
A methylation marker and kit technology, which can be used in biochemical equipment and methods, recombinant DNA technology, and microbial determination/examination, etc., can solve the problem of low sensitivity, invasiveness, and limited diagnostic ability of small lesions in urine exfoliative cytology. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
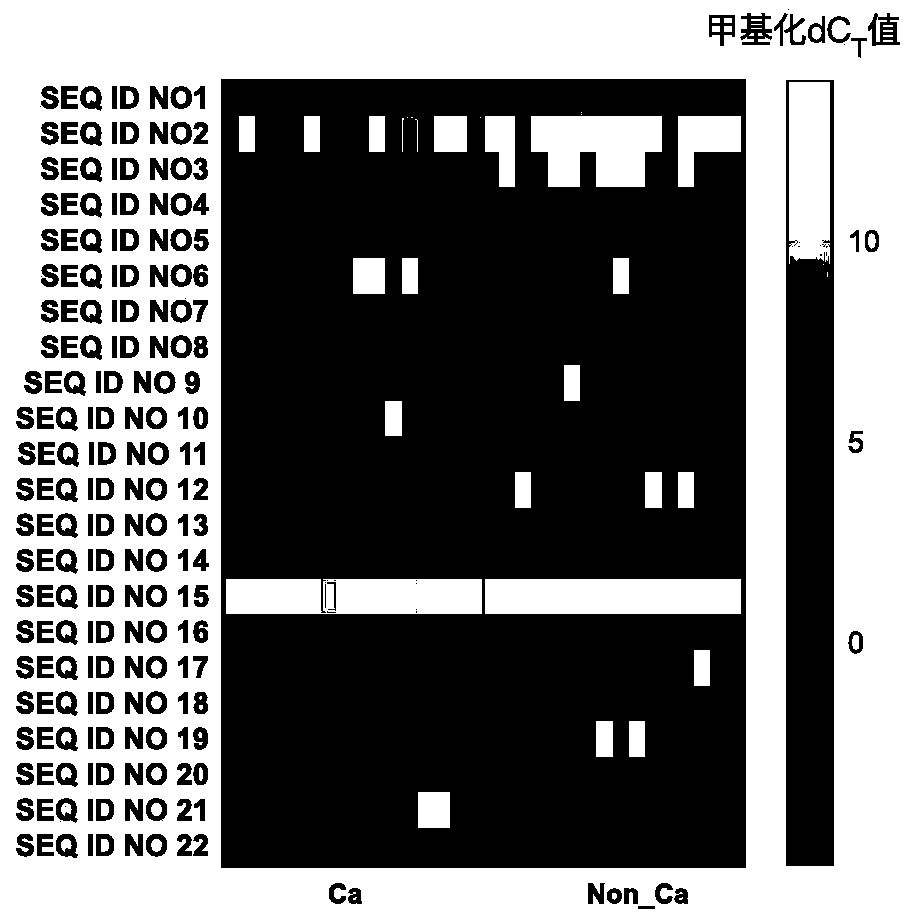
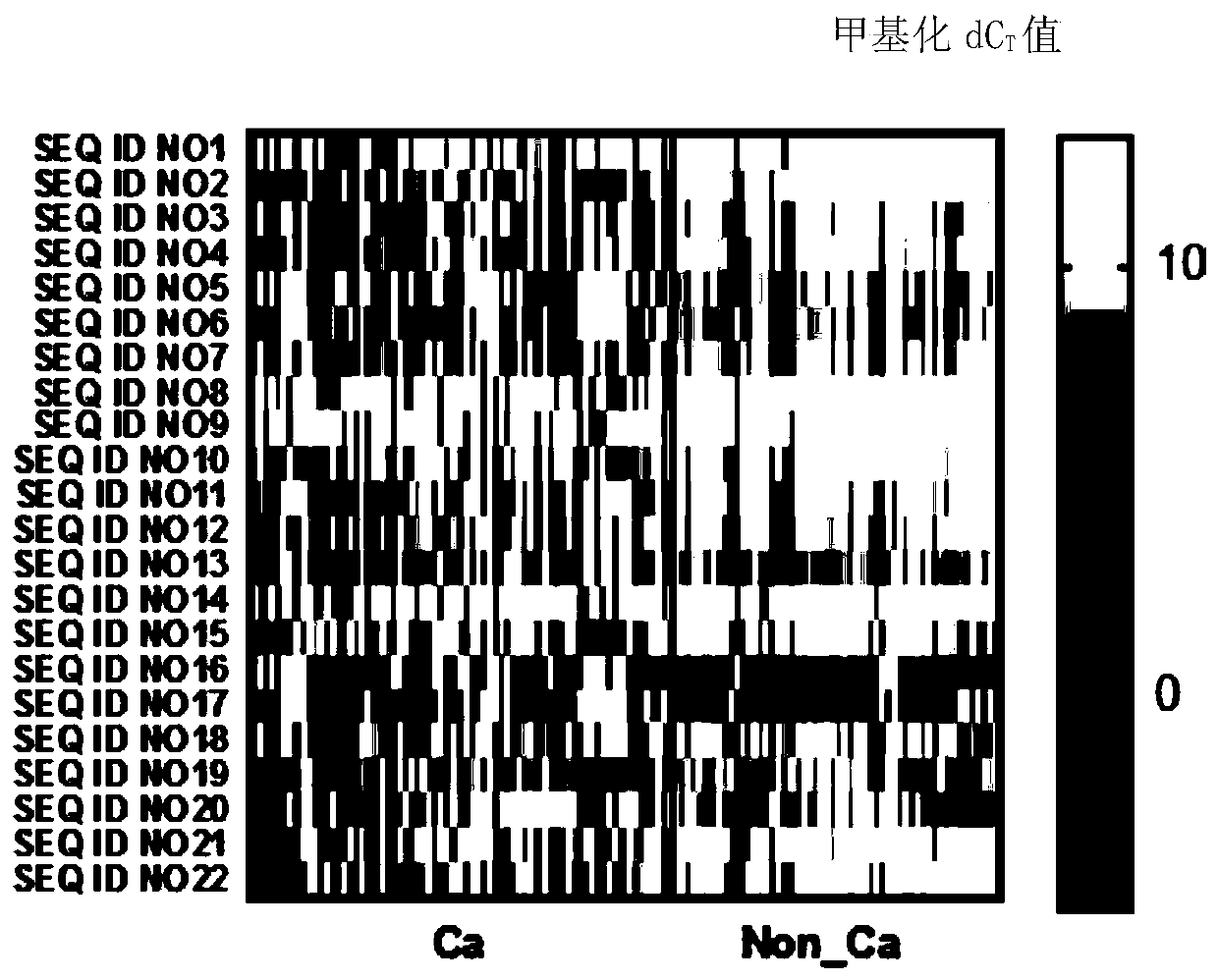
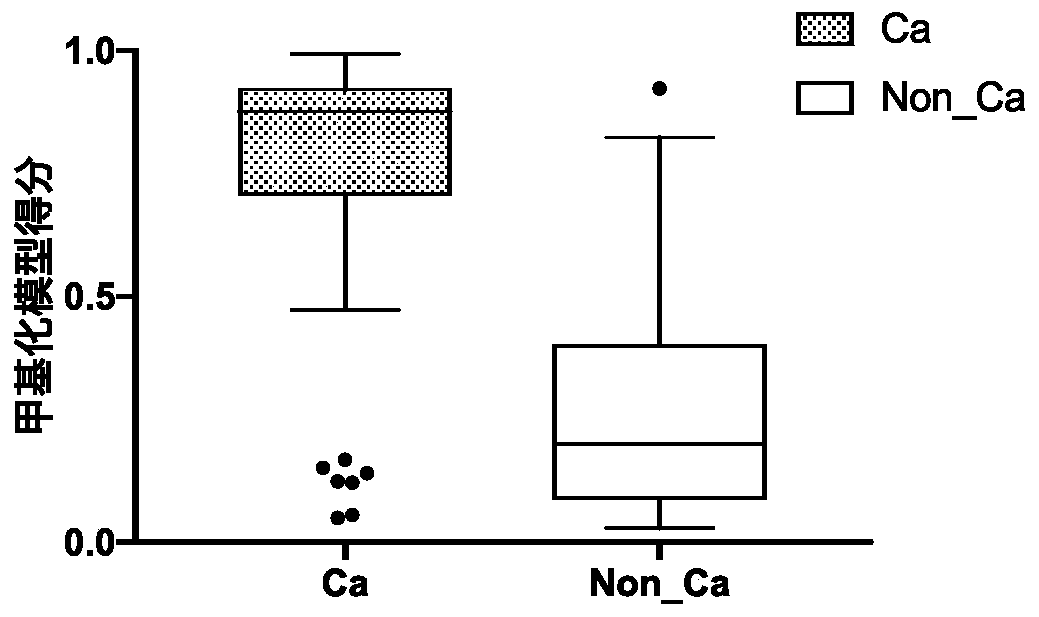
[0172] Co-methylation of methylated regions for bladder cancer detection, diagnosis, classification or prediction, treatment monitoring, prognosis or other evaluation of bladder cancer, including multiple methyl groups indicated by [CG] in the nucleic acid sequences listed in Table 1 Comethylation of methylation sites and comethylation of multiple methylation sites of nucleic acids that are completely complementary in sequence to the nucleic acids indicated by [CG] in Table 1.
[0173] Table 1 Co-methylation composition of DNA methylated regions
[0174]
[0175]
[0176]
Embodiment 2
[0178]A test kit for co-methylation of multiple methylated regions of bladder cancer for detection, diagnosis, classification or prediction, treatment monitoring, prognosis or other assessment of bladder cancer, comprising co-methylation of multiple methylated regions Specific primer pairs and probes, as shown in Table 2:
[0179] Table 2-1 Primer and probe sequence combinations for detection of co-methylation in 22 methylated regions 1
[0180]
[0181]
[0182]
[0183] Table 2-2 Primer and probe sequence combinations for detection of co-methylation in 22 methylated regions 2
[0184]
[0185]
[0186] Table 2-3 Primer and probe sequence combinations for detection of co-methylation in 22 methylated regions 3
[0187]
[0188]
[0189]
[0190] In practical applications, the corresponding primers and probes will be selected according to the combination of specific methylated regions.
[0191] Internal reference primers and probes:
[0192]
Embodiment 3
[0196] Co-methylation detection of 2-3 methylated regions by multiplex fluorescent quantitative PCR
[0197] Use commercial complete methylation (positive control) and non-methylation (negative control) standard (purchased from QIAGEN company) to carry out every 2-3 Co-methylation detection of methylated regions.
[0198] The specific process is as follows:
[0199] 1. DNA extraction
[0200] The extraction kit was purchased from QIAGEN, and was carried out according to the instructions of the kit.
[0201] 2. DNA bisulfite conversion
[0202] The DNA bisulfite conversion kit was purchased from Zymo, and was carried out according to the instructions of the kit.
[0203] 3. Multiplex PCR amplification
[0204] Using primer pairs for 22 methylated regions (SEQ ID NO.1-8), multiplex PCR was carried out in one reaction well (see Table 2 for the primer sequence), and the target sequence containing the target region was amplified, and the product size was in About 70-130bp.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com