Novel o-succinyl homoserine transferase mutant and method for producing o-succinyl homoserine using same
一种琥珀酰高丝氨酸、转移酶的技术,应用在O-琥珀酰高丝氨酸转移酶突变体领域,能够解决变异稳定性、低稳定性、恶化等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Example 1: Preparation of metX plasmid with O-acetyl homoserine transferase activity
[0097] To amplify the gene encoding O-acetyl homoserine transferase (MetX), based on a reporter sequence derived from wild-type (WT), a BamHI restriction enzyme site was inserted into the 300 bp) to the two ends of each primer (SEQ ID NO:5 and 6) for amplification of the stop region (located about 100 bp downstream of the stop codon).
[0098] Table 1
[0099] SEQ ID NO: Primer sequence (5'-3') 5 Primer 1 GGATCCCCTCGTTGTTCACCCAGCAACC 6 Primer 2 GGATCCCAAAGTCACAACTACTTATGTTAG
[0100] PCR was performed under the following conditions. After denaturation at 95°C for 5 minutes, the cycle (denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and polymerization at 72°C for 90 seconds) was repeated 30 times, and then polymerization was performed at 72°C for 7 minutes . As a result, a DNA fragment of 1546 bp was obtained as the coding r...
Embodiment 2
[0101] Embodiment 2: the preparation of the variant metX plasmid with O-succinyl homoserine transferase activity
[0102] A new metX mutation site was selected, and the 176th and 313th amino acids of the amino acid sequence of SEQ ID NO: 1 were respectively replaced with another amino acid.
[0103] More specifically, Q176N and L313R mutations were made. Using the pECCG117-metX WT plasmid prepared in Example 1 as a template, a primer pair (SEQ ID NO: 7 and 8) for the 176th mutation and a primer pair (SEQ ID NO: 8) for the 313rd mutation were designed. :9 and 10), to prepare a mutant vector in which the 176th amino acid of O-acetylhomoserine transferase was substituted by another amino acid and the 313th amino acid thereof was substituted by arginine.
[0104] Table 2
[0105] SEQ ID NO: Primer sequence (5'-3') 7 Primer 3 ACGCGCCAGCGCCTGGAACATCGGCATTCAATCCG 8 Primer 4 CGGATTGAATGCCGATGTTCCAGGCGCTGGCGCGT 9 Primer 5 GTAGATACCGATATTCGGTACC...
Embodiment 3
[0107] Example 3: Substrate specificity and activity of variant metX with O-succinyl homoserine transferase activity comparative test
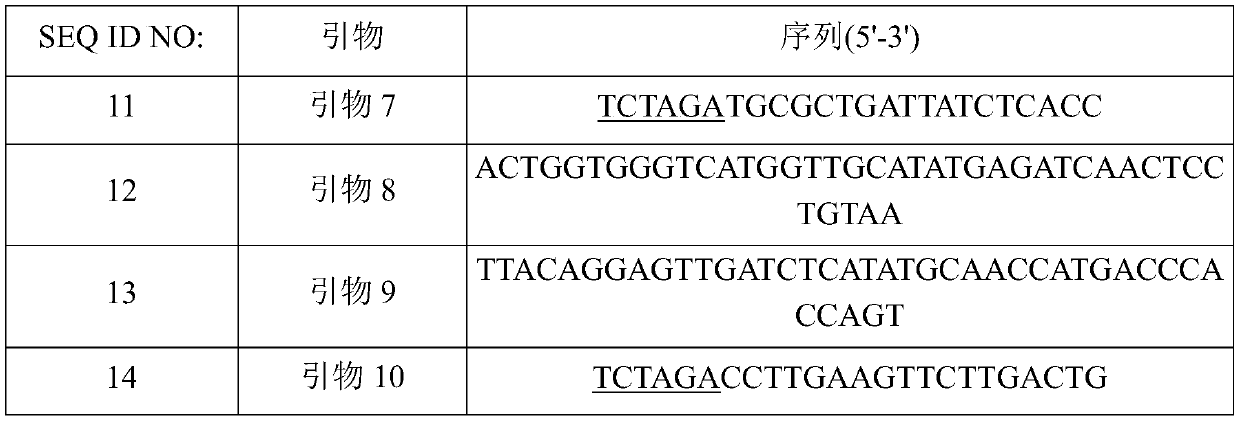
[0108] In order to compare the activity of mutated metX that produces excess O-succinyl homoserine, a strain in which homoserine was accumulated and utilization of the produced O-succinyl homoserine was absent was prepared. Deletion of metB gene encoding cystathionine gamma synthase in O-succinyl homoserine degradation pathway and metY deletion encoding O-acetyl homoserine (thiol)-lyase in O-succinyl homoserine degradation pathway Genetic strains. First, in order to delete the metB gene, based on the nucleotide sequence information of the WT-derived metB gene, a primer pair (SEQ ID NO: 11 and 12) for amplifying the 5' upstream region of the metB gene and a primer pair for amplifying the metB gene were designed. Primer pair for the 3' downstream region of the gene (SEQ ID NO: 13 and 14). An XbaI restriction enzyme site (underlined) was in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com