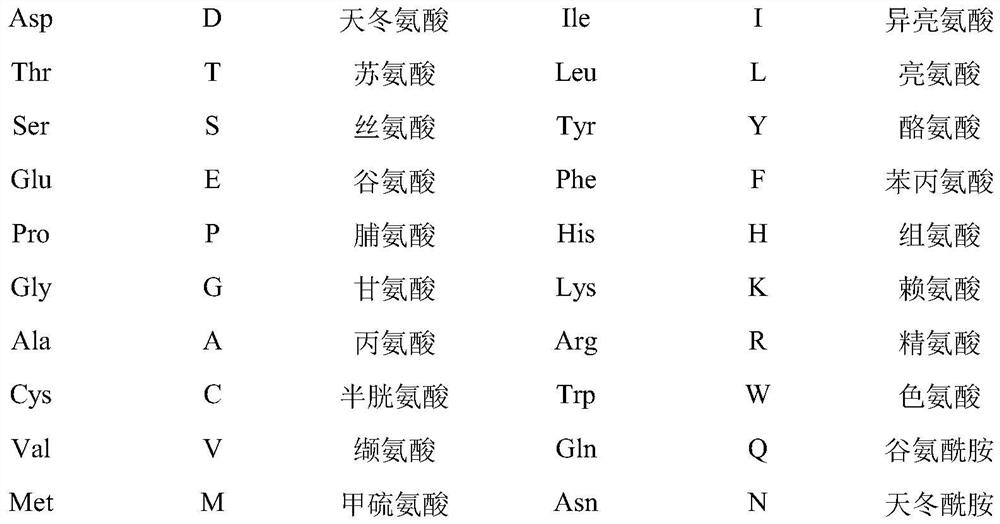
Short-chain dehydrogenase mutant and its application
A short-chain dehydrogenase and mutant technology, applied in the field of bioengineering, can solve the problems of low stereoselectivity, asymmetric reduction of carvone, single stereoselectivity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Construction of LfSDR1-M1 mutant gene by site-directed mutagenesis
[0030] Using the plasmid pET22b-LfSDR1 containing the short-chain dehydrogenase LfSDR1 gene as a template, the mutant gene was obtained by overlapping extension PCR.
[0031] First, the upstream and downstream primers of the LfSDR1 gene are as follows:
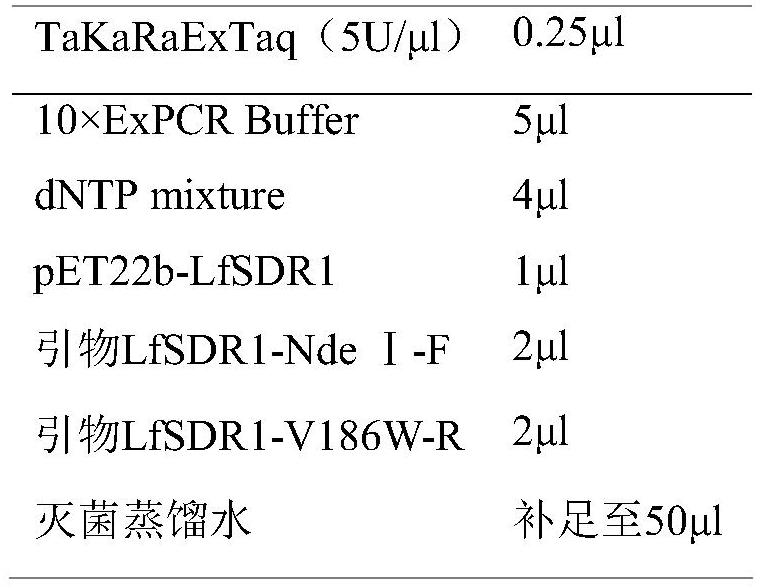
[0032] LfSDR1-NdeI-F: GGAATTCCATATGGGACAGTTTG
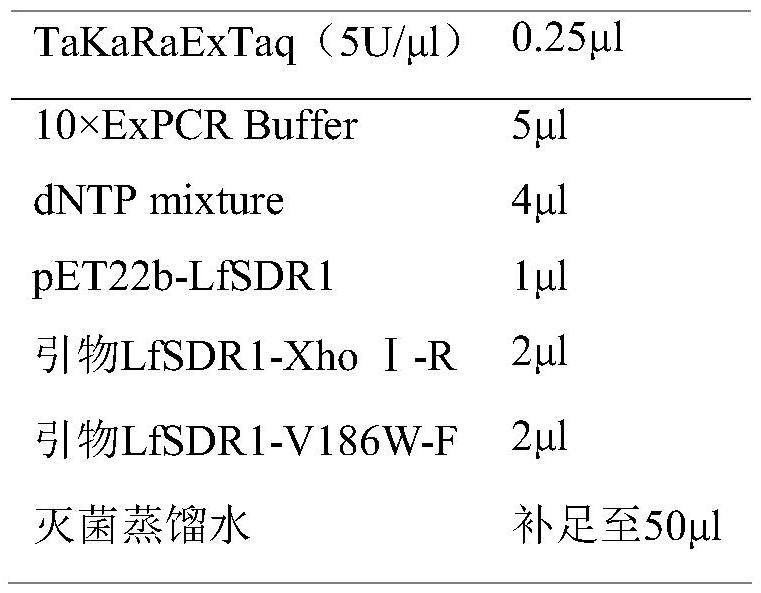
[0033] LfSDR1-XhoI-R: CCGCTCGAGTTGTGCCGTGTAGC
[0034] Then design mutation site primers as follows:
[0035] Mutant LfSDR1-M1
[0036] LfSDR1-V186W-F: TACCCTGGGTGGATTGCCACG
[0037] LfSDR1-V186W-R: CGTGGCAATCCACCCAGGGTA
[0038] The first step PCR reaction system is as follows:
[0039] PCR-a
[0040]
[0041] PCR-b
[0042]
[0043]Amplification program: 94°C: 10Min, (94°C: 30s, 45°C: 30s, 72°C: 30s) 30 cycles, 72°C, 10min. Two sets of PCR were used to obtain PCR fragment a and PCR fragment b respectively, and the two fragments were recovered with a common DNA product gel recove...
Embodiment 2
[0048] Example 2 Construction of LfSDR1-M2 mutant gene by site-directed mutagenesis
[0049] Using the plasmid pET22b-LfSDR1 containing the short-chain dehydrogenase LfSDR1 gene as a template, the mutant gene was obtained by overlapping extension PCR.
[0050] The upstream and downstream primers of the LfSDR1 gene are the same as in Example 1. LfSDR1 V186A / G92F / E141L / D146V / K206L
[0051] The primers for the mutation sites were designed as follows:
[0052] LfSDR1-G92F-F: GCCGGAATTTTTACTCCGCTG
[0053] LfSDR1-G92F-R: CAGCGGAGTAAAAATTCCGGC
[0054] LfSDR1-E141L-F: AGTTCGATCCTGGGGATGATC
[0055] LfSDR1-E141L-R: GATCATCCCCAGGATCGAACT
[0056] LfSDR1-D146V-F: ATGATCGGTGTGCCAACCGTT
[0057] LfSDR1-D146V-R: AACGGTTGGCACACCGATCAT
[0058] LfSDR1-V186A-F: TACCCTGGGGCAATTGCCACG
[0059] LfSDR1-V186A-R: CGTGGCAATTGCCCCAGGGTA
[0060] LfSDR1-K206L-F: TACATCGACCTGCACCCAATG
[0061] LfSDR1-K206L-R: CATTGGGTGCAGGTCGATGTA
[0062] Construction of LfSDR1-M2 mutants:
[0063] The fi...
Embodiment 3
[0082] Embodiment 3 construction mutant gene recombination plasmid
[0083] The mutant gene obtained by overlap extension PCR and the vector plasmid were subjected to double enzyme digestion, and the reaction system was as follows:
[0084]
[0085] After reacting at 37°C for 4-8 hours, use the ordinary DNA product gel recovery kit to recover the cleaved nucleic acid fragments; use T4 DNA ligase to ligate the double-digested mutant gene fragments with cohesive ends and the carrier plasmid fragments (16 °C reaction for 2-6h) to obtain recombinant plasmids pET22b-LfSDR1-M1 and pET22b-LfSDR1-M2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com