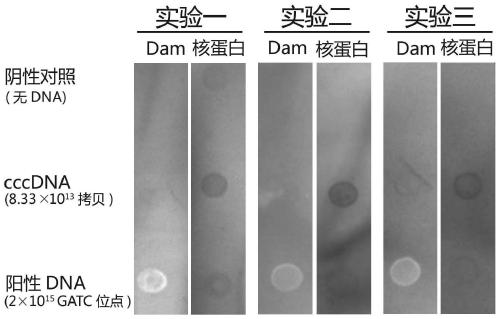
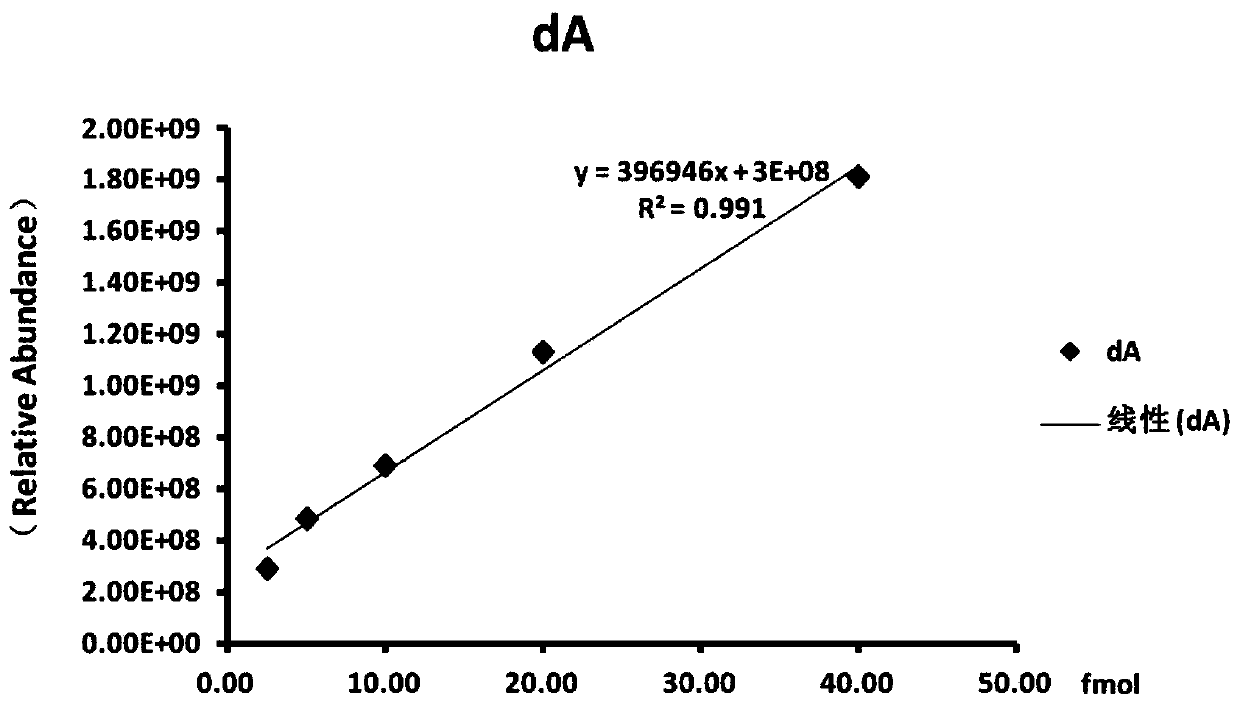
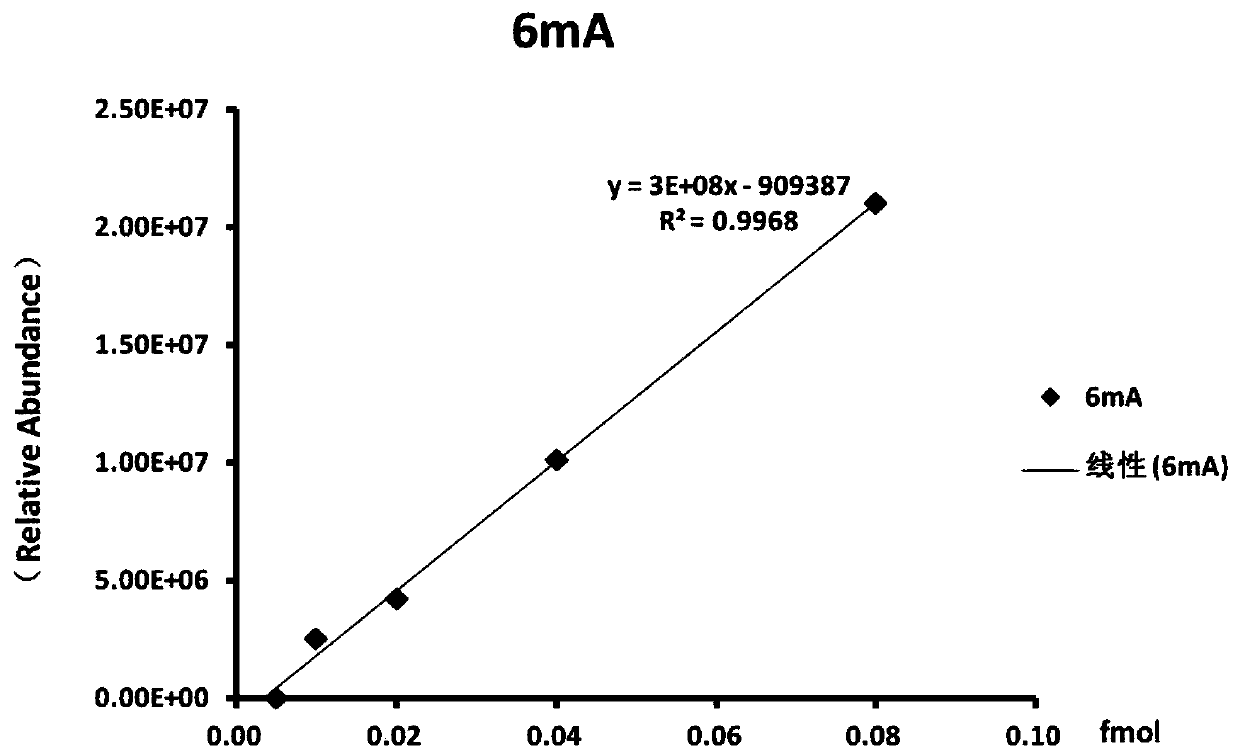
Method for modifying hepatitis B virus with N6-methyladenine in vitro
A technology of hepatitis B virus and methyl adenine, which is applied in the field of molecular biology, can solve the problems of low degree of methylation modification, inability to obtain a large amount of methylated HBV DNA, and has not yet been involved, and achieve the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] The method for in vitro N6-methyladenine modification of hepatitis B virus provided by the invention comprises the following steps:
[0021] 1. HBV viral genome cloning
[0022] The patient's serum DNA sample was provided by the Second Affiliated Hospital of Chongqing Medical University, China, and the patient number is 11061012. Using PrimeSTAR Max DNA polymerase, serum DNA template, front primer HBVF: 5'-CCGGAA AGCTTA TGCTCTTCTTTT TCACCT CTGCCT ARTCAT C-3', back primer HBVR: 5'-CCGGAG AGCTCA TGCTCT TCAAAAAGTTGC ATGGTG CTGGT-3' obtained by PCR amplification The full-length sequence of HBV with a length of 3.2kb. Then HBV was cloned into pEASY-Blunt vector to obtain pEASY-HBV positive plasmid. Through Sanger sequencing analysis and inspection, the present invention successfully obtains the full-length sequence of HBV DNA.
[0023] 2. In vitro circularization of HBV cccDNA
[0024] Using the successfully constructed pEASY-HBV plasmid of the present invention as a tem...
Embodiment 2
[0031] The method for in vitro N6-methyladenine modification of hepatitis B virus provided by the invention comprises the following steps:
[0032] 1. HBV viral genome cloning
[0033]The patient's serum DNA sample was provided by the Second Affiliated Hospital of Chongqing Medical University, China, and the patient number is 11061012. Using PrimeSTAR Max DNA polymerase, serum DNA template, front primer HBVF: 5'-CCGGAA AGCTTA TGCTCTTCTTTT TCACCT CTGCCT ARTCAT C-3', back primer HBVR: 5'-CCGGAG AGCTCA TGCTCT TCAAAAAGTTGC ATGGTG CTGGT-3' obtained by PCR amplification The full-length sequence of HBV with a length of 3.2kb. Then HBV was cloned into pEASY-Blunt vector to obtain pEASY-HBV positive plasmid. Through Sanger sequencing analysis and inspection, the present invention successfully obtains the full-length sequence of HBV DNA.
[0034] 2. In vitro circularization of HBV cccDNA
[0035] Using the successfully constructed pEASY-HBV plasmid of the present invention as a temp...
Embodiment 3
[0042] The method for in vitro N6-methyladenine modification of hepatitis B virus provided by the invention comprises the following steps:
[0043] 1. HBV viral genome cloning
[0044] The patient's serum DNA sample was provided by the Second Affiliated Hospital of Chongqing Medical University, China, and the patient number is 11061012. Using PrimeSTAR Max DNA polymerase, serum DNA template, front primer HBVF: 5'-CCGGAA AGCTTA TGCTCTTCTTTT TCACCT CTGCCT ARTCAT C-3', back primer HBVR: 5'-CCGGAG AGCTCA TGCTCT TCAAAAAGTTGC ATGGTG CTGGT-3' obtained by PCR amplification The full-length sequence of HBV with a length of 3.2kb. Then HBV was cloned into pEASY-Blunt vector to obtain pEASY-HBV positive plasmid. Through Sanger sequencing analysis and inspection, the present invention successfully obtains the full-length sequence of HBV DNA.
[0045] 2. In vitro circularization of HBV cccDNA
[0046] Using the pEASY-HBV plasmid successfully constructed in the present invention as a tem...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com