Preparation and application of aptamer resisting potassium perfluorooctane sulfonate
A technology of potassium perfluorooctane sulfonate and nucleic acid aptamer, which is applied in the direction of biochemical equipment and methods, instruments, and analytical materials, and can solve problems such as expensive instruments and complicated operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] 1) Design a random single-stranded library and combine it with downstream primers:
[0038] 5'-GCGCATACGAGCTTGTTCAATA (SEQ ID NO.05)
[0039] -N40-TGATAGTAAGTGCAATCTAGCC (SEQ ID NO.06)-3' and biotin-modified downstream primer Biotin-PR: Biotin-C6-GGCTAGATTGCACTTACTATCA-3' (SEQ ID NO.07), molar ratio 1:1~1:2 , optimize the ratio of 1:1.5 into a 200μL PCR tube, add an appropriate amount of binding buffer (5 × binding buffer containing 125mM Tris-HCl, 500mM NaCl, 125mMKCl, 50mM MgCl 2 , 5% DMSO, pH7.5) diluted to 1×bindingbuffer in a PCR tube to 200μL and mixed. Then put it in a PCR instrument: 95°C for 10 minutes, 4°C for 10 minutes, and slowly return to room temperature for annealing;
[0040] 2) Take 10 μL of streptavidin magnetic bead suspension (Thermo Fisher, USA) in a 1.5 mL centrifuge tube, add 200 μL of 1×binding buffer to wash, and let stand on the magnetic stand for 1 min, until no magnetic beads are suspended in the supernatant, Remove the supernatant and re...
Embodiment 2
[0056] 1) Pretreatment of nitrocellulose membrane: Use scissors to cut a few pieces of nitrocellulose membrane (only one sample) with a size of about 5 mm × 5 mm, put them on clean PE gloves that have been marked in advance, and use a pencil to mark the surface of the membrane. Mark the upper right corner as the site for tweezers to prevent large-scale contamination of the membrane, and use a 1mL syringe needle to position in the center of the membrane;
[0057] 2) Target immobilization: take 2.5 μL of sample (experimental group: target solution; primer control group: target solution; bindingbuffer control group: binding buffer) and place it in the center of the marked area of the syringe, and apply additional samples after air-drying for the second time Leave it at room temperature for 1.5h;
[0058] 3) Membrane washing: Place the nitrocellulose membrane immobilized with the target in a 24-well plate (one membrane per well), add 1 mL washing buffer (binding buffer containin...
Embodiment 3
[0073] High-throughput sequencing and result analysis:
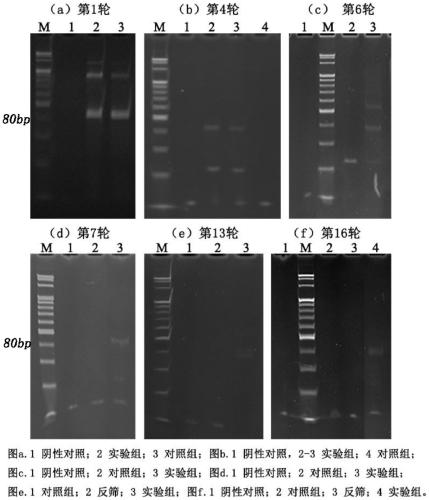
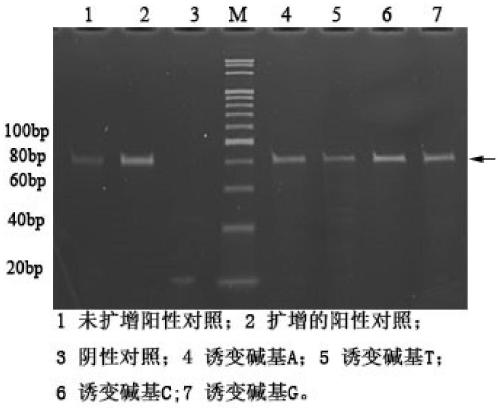
[0074] 1) In order to avoid errors in high-throughput sequencing caused by contamination, the samples were subjected to PCR amplification and electrophoresis before sequencing ( Figure 5 );
[0075] 2) The amplified sample was subjected to high-throughput sequencing (llumina, MiSeq2x300bp.), and analyzed from three aspects: sequence homology, secondary structure diversity and complexity, and three-dimensional structure molecular docking, and the sequence obtained from high-throughput sequencing 4 possible candidate sequences were selected from
[0076] PS1:
[0077] GCGCATACGAGCTTGTTCAATATGAGGCTTGTCGAATAACGTGTCTAATCGAGTCCCCGCCGTTGATAGTAAGTGCAATCTAGCC( Image 6 , SEQ ID NO.01)
[0078] PS36:
[0079] GCGCATACGAGCTTGTTCAATAATCGTGCTGTGCGCATACGAGCTTGTTCAATAGTGGTTGATAGTAAGTGCAATCTAGCC( Figure 7 , SEQ ID NO.02)
[0080] PS39:
[0081] GCGCATACGAGCTTGTTCAATATGAGGCTCGTCGAATAACGTGTCTAATCGAGTCCCCGCCGTTGATAGTAAGTGCAATCTAG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com