Probe library for detecting fusion gene of neurotrophin receptor kinase (NTRK) gene family, reagent, kit and application
A technology that integrates genes and gene families. It is applied in the direction of DNA/RNA fragments, recombinant DNA technology, and microbial measurement/inspection. It can solve the problems of missed detection at the RNA level, false positives, and the inability to know the gene sequence.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The following examples are intended to specifically illustrate the probes, detection reagents, applications, kits, and corresponding detection methods involved in the present invention.
[0047] In this example, a specific probe library capture and reversible end-stop sequencing technology was used to extract deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) from formalin-fixed paraffin-embedded solid tumor tissue (FFPE) samples Purify, then construct a DNA and cDNA sample sequencing library, and use a specific probe library to capture and enrich the target area of the library. The captured library is sequenced through high-throughput, so as to realize the double-layered DNA and RNA of the NTRK gene family fusion gene One-time detection of multiple fusion forms.
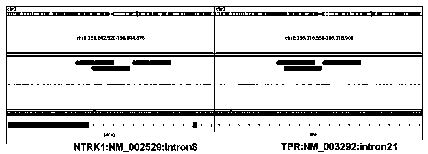
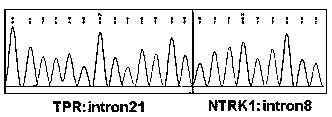
[0048] The probe library involved in this embodiment is based on known fusion partner genes, such as AFAP1, AGBL4, ARHGEF2, BCAN, BCR, BTBD1, CD74, CHTOP, EML4, ETV6, IRF2BP2, LMNA, MPRIP, MYO5A, NACC2, NFAS...
Embodiment 2
[0270] This example is to illustrate the optimization experiment of the number of cycles of library amplification processing.
[0271] The input amount (sample addition amount) can affect the success rate of the test. Within the sample addition standard established by the laboratory (50~1000ng), when it is higher or lower than the concentration range, the test success rate and test consistency are both Various degrees of decline. Therefore, in the library construction process, it is necessary to determine the appropriate sample amount range. Therefore: for the DNA nucleic acid extracted from the sample to be tested, in Example 1, the total amount of nucleic acid extracted is at least 50-500ng, of which 50ng is the smallest The dosage is the minimum input amount of gDNA that can be detected in this embodiment; similarly, for RNA, the total amount of nucleic acid extracted is at least 100 ng, which is the minimum input amount of RNA that can be detected in this embodiment.
[0272] ...
Embodiment 3
[0299] This example uses the probe library provided in Example 1 and the corresponding kit, and uses the detection method of Example 1 to test different samples to verify the specificity of the probe library provided in Example 1. The test results are shown in the table 29 shown.
[0300]
[0301] In Table 29:
[0302] (1) Nos. 1-3 are other fusion samples that are not known to be NTRK family fusion genes. According to the results, it can be seen that using the probe and method of Example 1, the test results show that NTRK is negative, that is, no NTRK family is detected Fusion genes, which shows that the probe library of Example 1 cannot capture non-NTRK family fusion genes;
[0303] (2) The sample No. 4 is a bacterium that does not have NTRK family fusion genes. The test results show that there is no interference from non-human sources, indicating that the probe library of Example 1 does not cross-react with non-human nucleic acids;
[0304] (3) The samples with serial numbers 5-8 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| surface area | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com