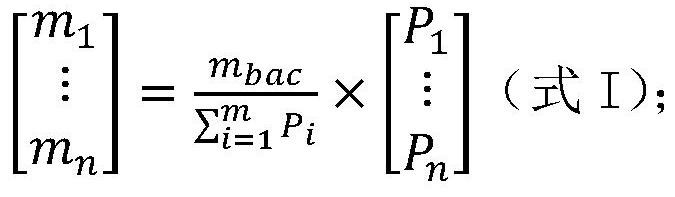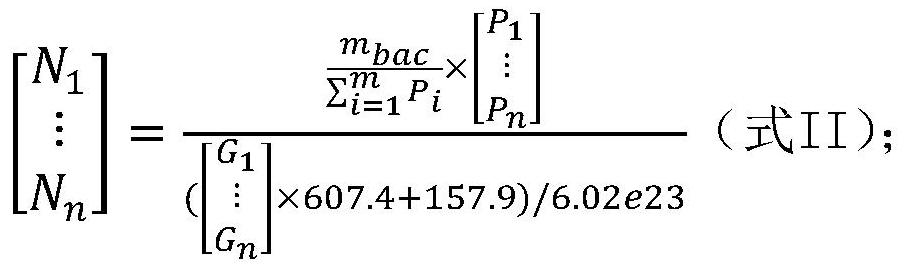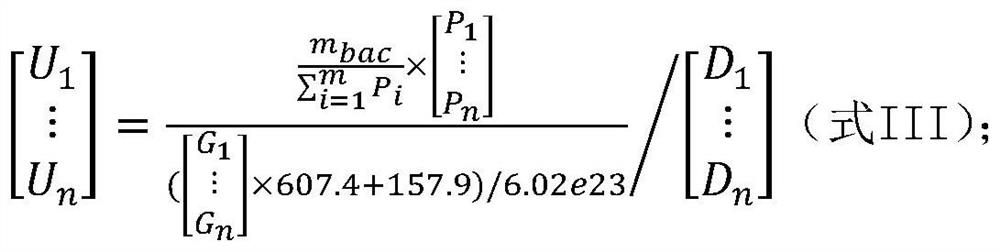A method for detecting the genome dna copy number of each microbial species in a sample to be tested
A genome and microbial technology, applied in the field of microorganisms, can solve problems such as time-consuming and labor-intensive, inability to achieve high-throughput detection, and achieve high-accuracy results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0141] Embodiment 1, detect total bacterial genome DNA content m in the sample to be tested bac , the genome content of each microbial species in the sample to be tested m i , genome copy number N i and the number of cells (number of viral particles) U i the establishment of the method
[0142] The inventor of the present invention has established the detection of the total bacterial genome DNA content m in the sample to be tested through a large number of experiments. bac , the genome content of each microbial species in the sample to be tested m i , genome copy number N i and the number of cells (number of viral particles) U i method, the specific steps are as follows:
[0143] 1. UCP DNA Micro Kit was used to extract the total DNA of the sample to be tested.
[0144] 2. After completing step 1, take the total DNA of the sample to be tested, and use the human pathogenic nucleic acid detection kit to construct a metagenomic next-generation sequencing library of the sam...
Embodiment 2
[0156] Embodiment 2, using the method established in Example 1 to detect the total bacterial genome DNA content in bronchoalveolar lavage fluid m bac , genome content of each microbial species in bronchoalveolar lavage fluid m i , genome copy number N i and the number of cells (number of viral particles) U i
[0157] 1. Obtaining total DNA from bronchoalveolar lavage fluid
[0158] Providers of bronchoalveolar lavage fluid in this example gave informed consent.
[0159] Take 600 μL of bronchoalveolar lavage fluid (ie sample), and use UCP DNA Micro Kit to extract total DNA to obtain the total DNA of bronchoalveolar lavage fluid.
[0160] A total of 20 μL of eluate was collected. After testing, the concentration of total DNA in the bronchoalveolar lavage fluid was 5.3 ng / μL.
[0161] 2. Construction of metagenomic next-generation sequencing library of bronchoalveolar lavage fluid
[0162] The total DNA of bronchoalveolar lavage fluid was collected, and a metagenomic next-...
Embodiment 3
[0204] The accuracy experiment of the method that embodiment 3, embodiment 1 establish
[0205] 1. Preparation of Test Articles
[0206] Staphylococcus epidermidis, Acinetobacter baumannii, Candida albicans and Epstein-Barr virus are all products of ATCC, and the product numbers are 12228, 19606, 14053 and VR-1492 respectively.
[0207] 1. UCP DNA Micro Kit was used to extract the total DNA of Staphylococcus epidermidis, Acinetobacter baumannii, Candida albicans and Epstein-Barr virus.
[0208] 2. Using the Qubit dsDNA HS Assay Kit, refer to the Qubit instrument manual to detect the mass concentration of the total DNA of Staphylococcus epidermidis, the total DNA of Acinetobacter baumannii, the total DNA of Candida albicans, the total DNA of Epstein-Barr virus and the host nucleic acid. The host nucleic acid is human nucleic acid.
[0209] The results showed that the total DNA concentration of Staphylococcus epidermidis was 2.96ng / μL, the total DNA concentration of Acinetobac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com