Method and system for detecting degree of tumor heterogeneity
A heterogeneity, tumor technology, applied in the field of bioinformatics, can solve the problem of inability to detect the degree of tumor heterogeneity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
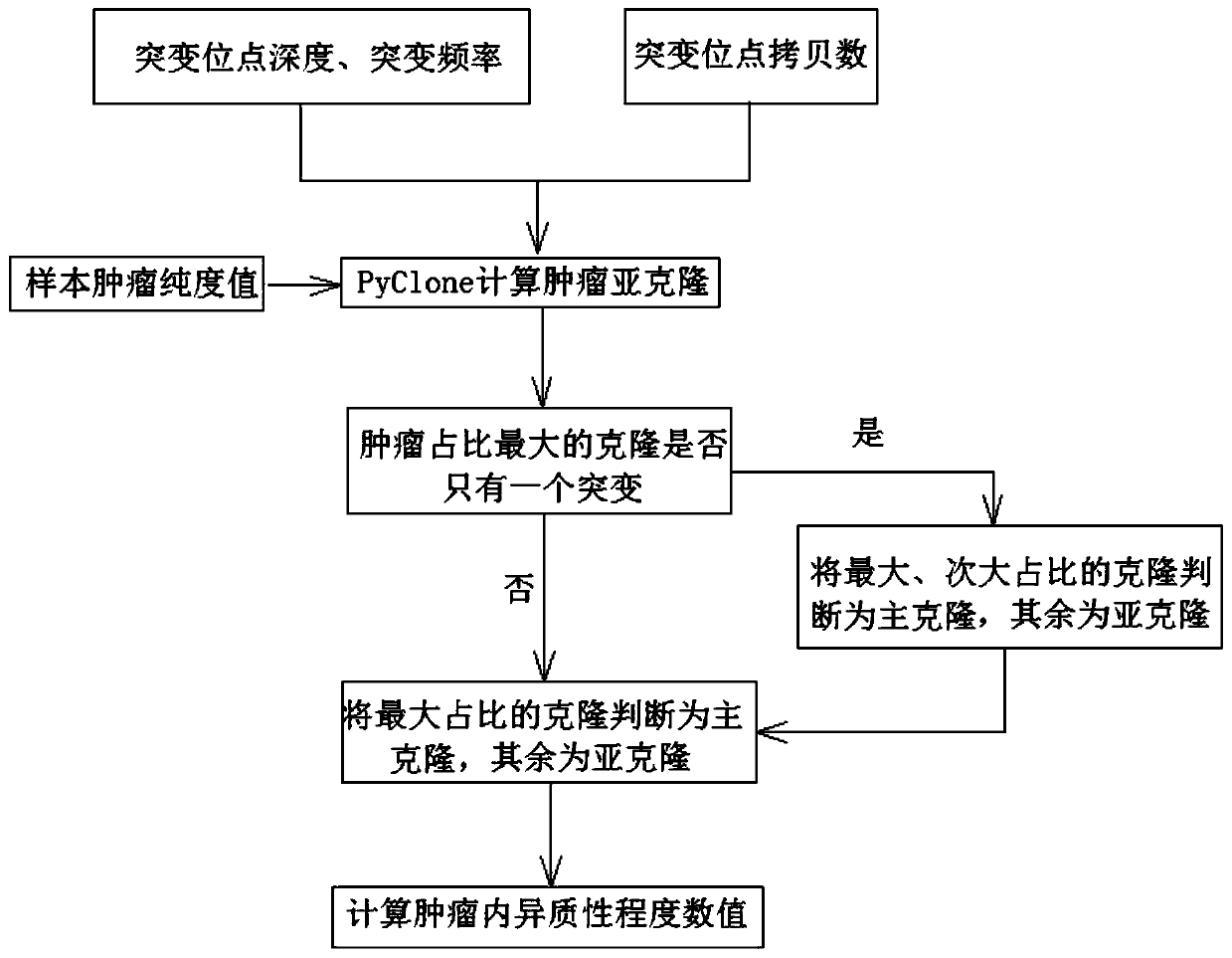
[0118] In this embodiment, the samples used are a batch of tumor tissue samples and control blood samples from a lung cancer immunotherapy cohort of 69 patients in total, and the control blood samples are specifically white blood cell samples separated from peripheral blood. The sampling method of the tumor tissue sample in this embodiment is a formalin-fixed paraffin-embedded (FFPE) sample prepared by sampling the pathological puncture of each patient's lung lesion at a single point, and the sampling unit is the Cancer Prevention and Control Center of Sun Yat-sen University.
[0119] The specific steps of detecting tumor heterogeneity of paired samples in this embodiment are as follows:
[0120] 1) Obtain tumor tissue samples and control blood samples, and perform whole exome sequencing. This sequencing is provided by Nanjing Shihe Gene Biotechnology Co., Ltd. After obtaining the sequencing sequence data, use the variation detection software VarScan (v2.4.1) to detect the var...
Embodiment 2
[0137] In this embodiment, the samples used are a batch of tumor tissue samples and control blood samples from a nasopharyngeal carcinoma immunotherapy cohort of a total of 56 patients, and the control blood samples are specifically white blood cell samples separated from peripheral blood. The sampling method of the tumor tissue sample in this embodiment is sampling at a single point in each patient's lung lesion. The sampling method of the tumor tissue sample in this embodiment is the pathological puncture single-point sampling of the lung lesions of each patient and the prepared formalin-fixed paraffin-embedded (FFPE) sample, and the sampling unit is the Cancer Prevention and Control Center of Sun Yat-sen University.
[0138] The specific steps of detecting tumor heterogeneity of paired samples in this embodiment are as follows:
[0139] 1) Obtain tumor tissue samples and control blood samples for whole exome sequencing, which is provided by Nanjing Shihe Gene Biotechnology ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com