Application of target gene in shellfish larva metamorphosis development process researched by RNA interference technology
A technology of RNA interference and technical research, which is applied to the application field of genes in the metamorphic development of shellfish larvae, can solve the problems of small individual shellfish larvae and cannot use injection methods, etc. Eliminate the effect of genetic interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
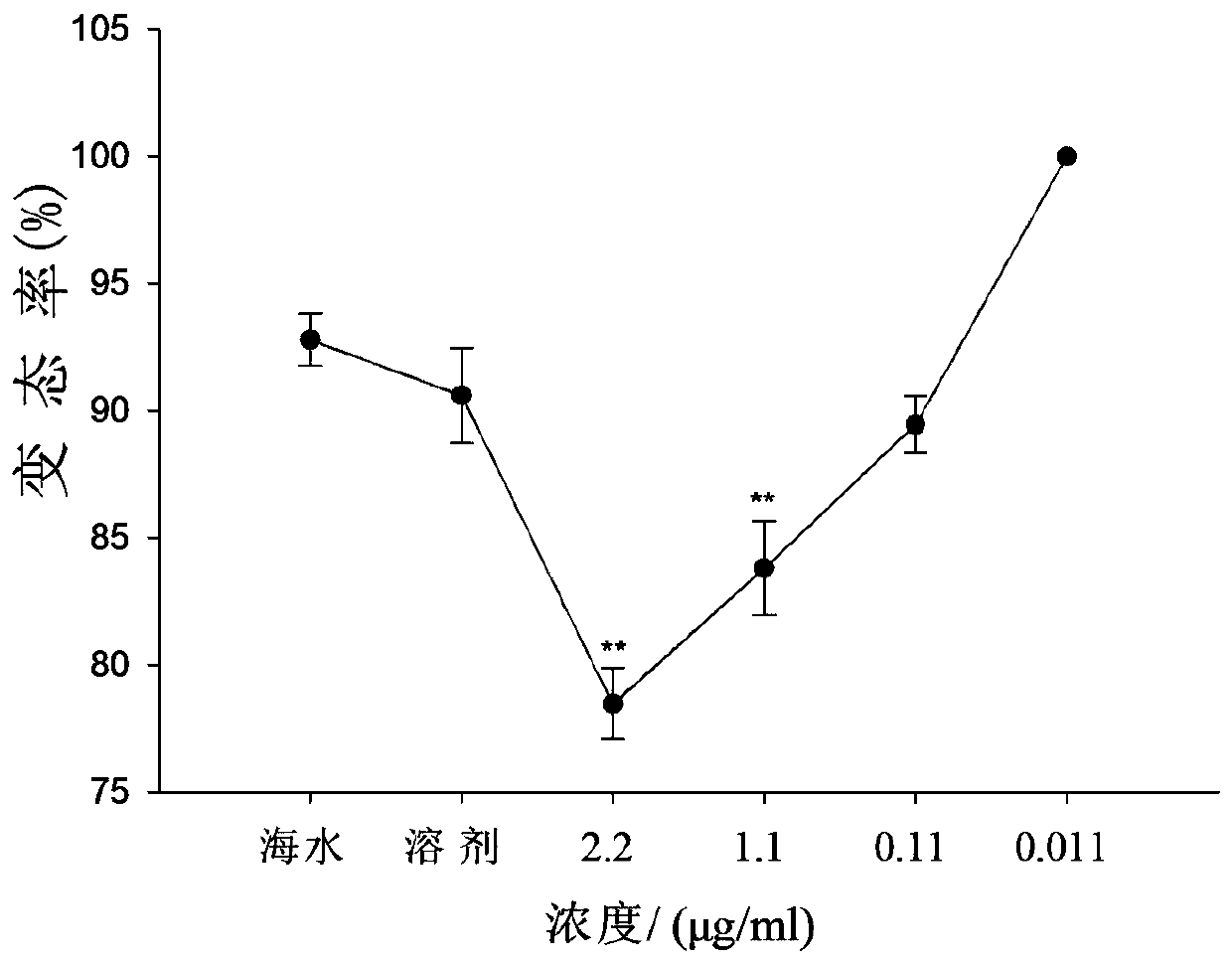
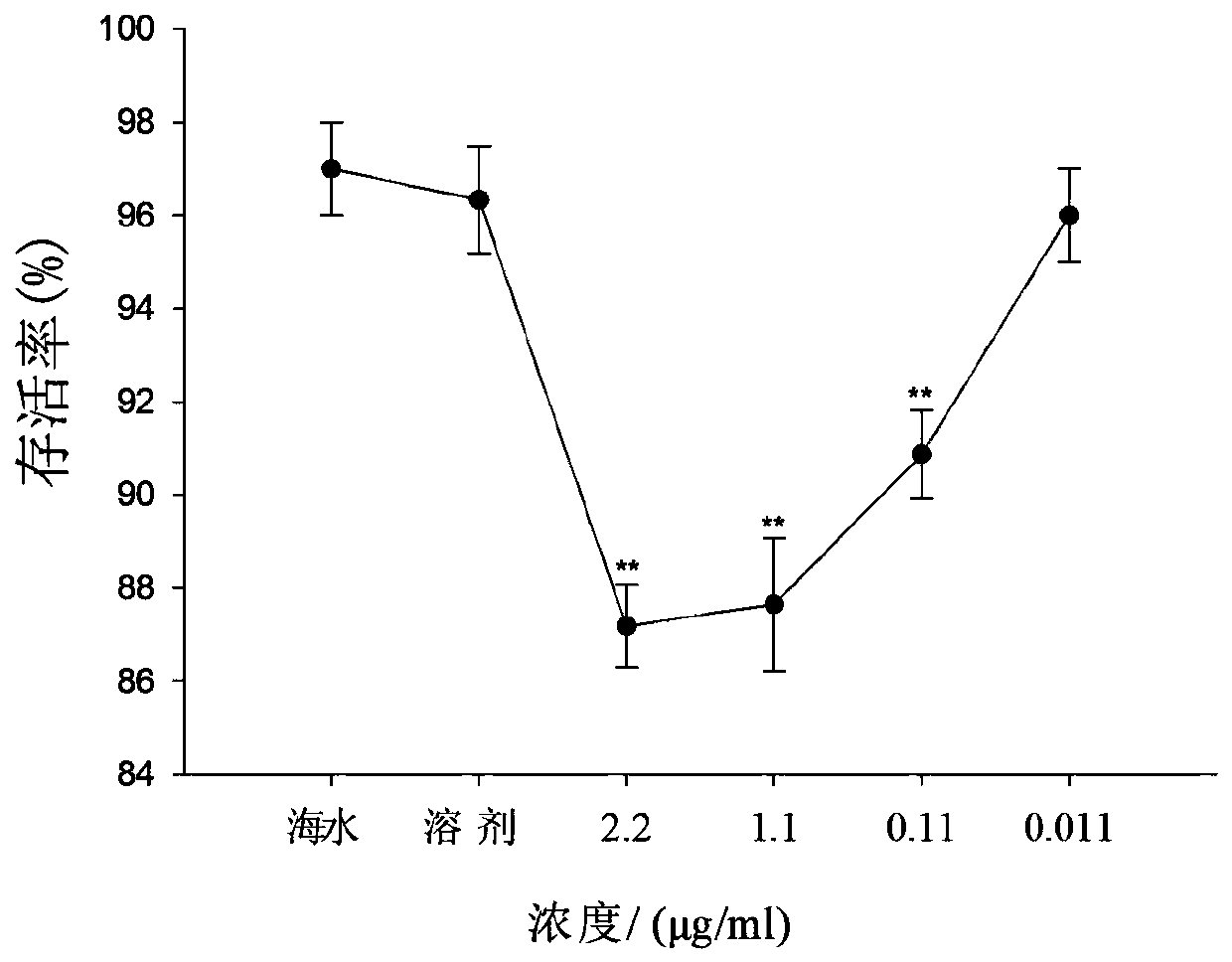
[0061] The phenotype observation experiment used a six-hole plate, with 100 razor clam larvae in each well, and 10ml of seawater (containing 2.5×10 5 of golden algae and 2.5 x 10 5 Chaetoceros), different concentrations of siRNA or DEPC reagent. The metamorphosis rate was observed every 4 hours. When the seawater group was completely metamorphosed, the metamorphosis rate and survival rate of each group were calculated. Finally, from the phenotype data, compared with the blank group and DEPC group, the metamorphosis rate of larvae in all siRNA groups decreased with the increase of siRNA concentration. It can be seen from the phenotype that during the development period of razor clam larvae, siRNA made the metamorphosis rate of razor clam ( figure 1 ) and survival rate ( figure 2 ) was significantly suppressed. It can be inferred that in the developmental stage of razor clam larvae, when siRNA silences the target gene-dopamine β-hydroxylase, its activity is inhibited, the n...
Embodiment 2
[0063] In this example, razor clam larvae from the shell top stage to the creeping stage were selected to collect samples at different time points after the disturbance. A total of 5 time points were designed, namely: 0h, 4h, 8h, 12h and 24h, and the corresponding DEPC groups were designed, and each group had three repetitions. 500ml volume beakers were used in the experiment, each containing 200ml seawater and razor clam larvae. Collect razor clam larvae at each time point, collect razor clam larvae with an enrichment net, pipette the larvae into a 1.5ml centrifuge tube, centrifuge and remove excess seawater, add RNA store and store in a -20°C refrigerator for later use Detection of gene expression.
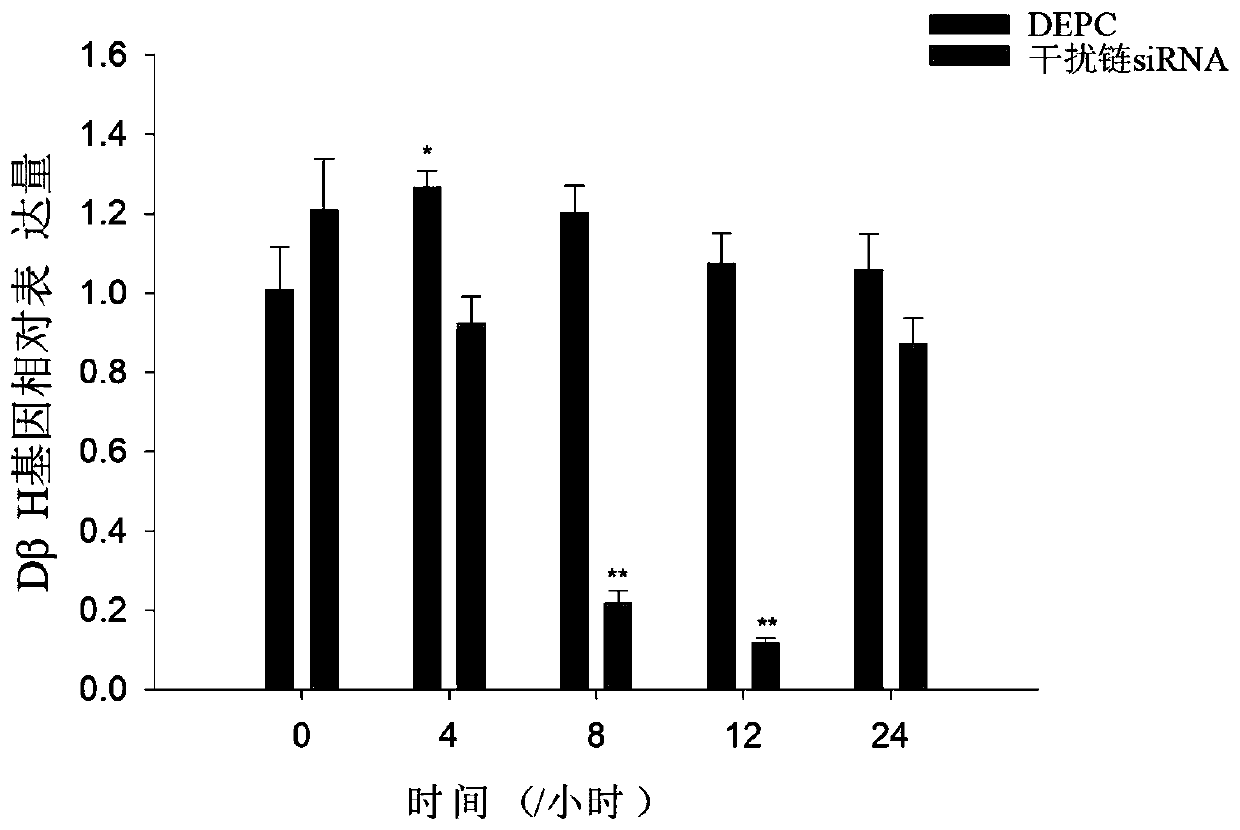
[0064] Finally, from the results of gene expression: during the development of razor clam larvae from the top of the shell to the creeping stage, the inhibitory effect of siRNA on the target gene was detected at different soaking times ( image 3 ), thereby indicating the sile...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com