Preparation method and application of Cas9-RNAi RNP with efficient homologous directional repair activity
An active and efficient technology, applied in the field of recombinant proteins, can solve problems such as ectopic rearrangement of chromatin, inability to repair cells in time, and cancerous cell death, etc., and achieve the effects of efficient repair, cost reduction and efficiency improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
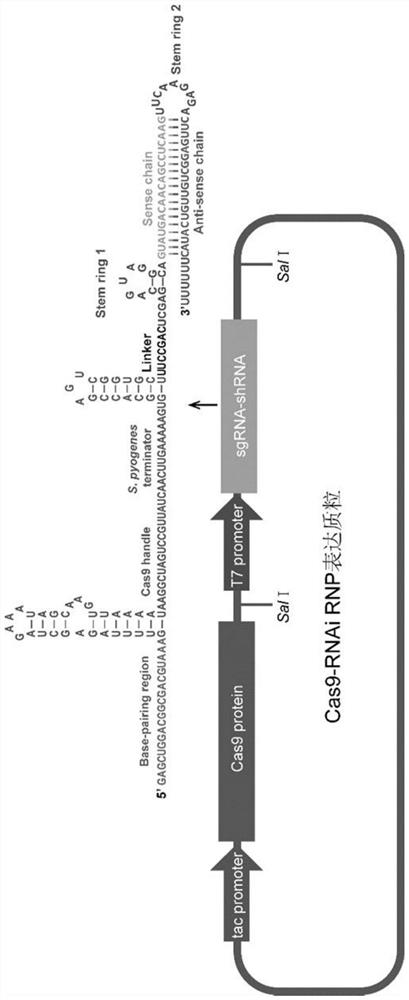
[0045] Example 1 Construction of Cas9-RNAi RNP expression plasmid
[0046] 1. The Ptac-cas9-T7sgRNA plasmid stored in our laboratory is the backbone of the vector, which is digested with the restriction enzyme SalI. The SpCas9 gene sequence already exists on the plasmid, and the sgRNA-shRNA sequence only needs to be connected after the restriction site of SalI.
[0047] 2. Design sgRNA-shRNA sequences targeting BFP gene and interfering Ligase 4 gene with a total length of 210nt. Five pairs of primers were synthesized and the sgRNA-shRNA fragments were constructed by annealing the homology arms, and then PCR amplification was carried out. When replacing the sgRNA sequence and shRNA sequence, only 2 pairs of primers need to be redesigned, and the other 3 pairs of primers remain unchanged. The mixed primer in overlapping PCR is to mix and dilute other primers except the first and last primers by 10 times. The cycle number of overlapping PCR is about 10. The product is used as a...
Embodiment 2
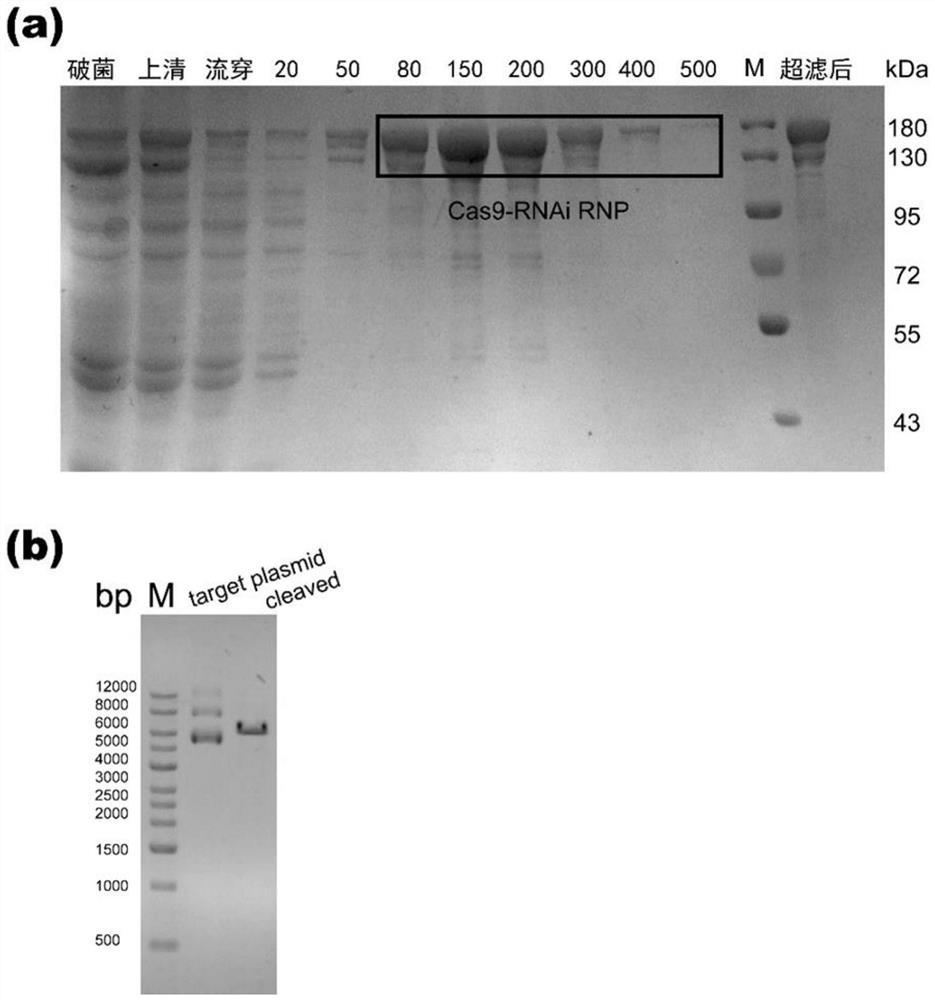
[0073] Example 2 Expression purification and in vitro activity test of Cas9-RNAi RNP
[0074] like figure 1 As shown, the Cas9-RNAi RNP expression plasmid constructed in Example 1 was transformed into E. coli expression strain HT115 DE3 competent cells, and the target expression strains were obtained by screening with ampicillin and tetracycline double antibodies. Introduce this strain into the liquid medium containing double antibody, cultivate overnight in a shaker at 37°C 220rpm, then transfer to 1L liquid medium containing corresponding antibiotics at 1:100, and continue to culture at 37°C 220rpm to OD600 =0.6~0.8. After adding IPTG with a final concentration of 1 mM, it was transferred to a shaker at 18°C and 220 rpm for induction for 16-18 hours. After induction, the cells were collected and disrupted, and a large number of Cas9-RNAi RNPs were rapidly prepared by one-step Ni-bead affinity purification and analyzed by SDS-PAGE electrophoresis. The purification result...
Embodiment 3
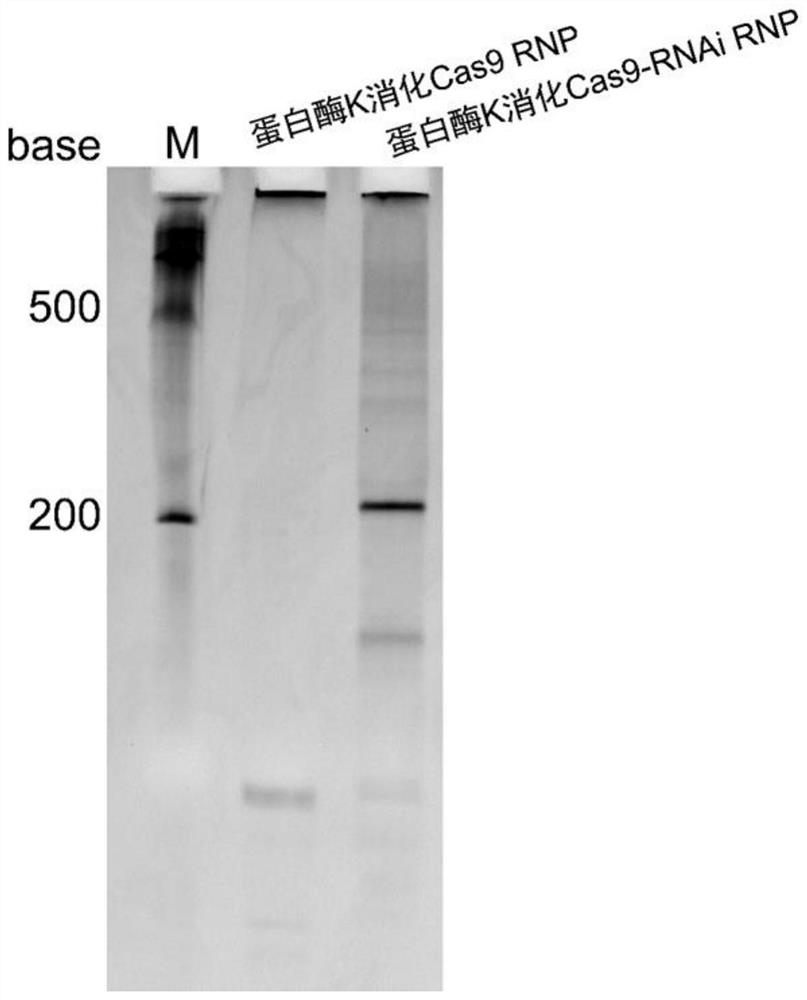
[0076] Example 3 Detection of the integrity of sgRNA-shRNA in Cas9-RNAi RNP
[0077] In order to detect whether the protein obtained in Example 2 contains complete sgRNA-shRNA, the purified Cas9-RNAi RNP was treated with proteinase K, and the results of polyacrylamide gel nucleic acid electrophoresis were as follows: image 3 shown. The theoretical size of sgRNA-shRNA is about 210nt, and the electrophoresis results are consistent with expectations.
[0078] The results of this example show that the constructed Cas9-RNAi RNP expression plasmid can be correctly expressed when transformed into E. coli HT115(DE3), and the self-assembled SpCas9 RNP containing sgRNA-shRNA structure in E. coli can be obtained by Ni-bead affinity purification in one step.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com