Synechocystis genetically engineered bacterium for biosynthesizing inositol as well as construction method and application thereof
A genetically engineered bacteria and biosynthetic technology, applied in the field of Synechocystis genetically engineered bacteria and its construction for biosynthesizing inositol
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Construction of strain WT-INO1SS:
[0038] (1) Using the whole genome of Saccharomyces cerevisiae BY4741 as a template, using the nucleotide sequences shown in SEQ ID NO.10 and SEQ ID NO.11 in the sequence listing as upstream and downstream primers, amplified to obtain inositol- 1-monophosphatase gene INO1, the nucleotide sequence of the INO1 gene is shown in SEQ ID NO.1;
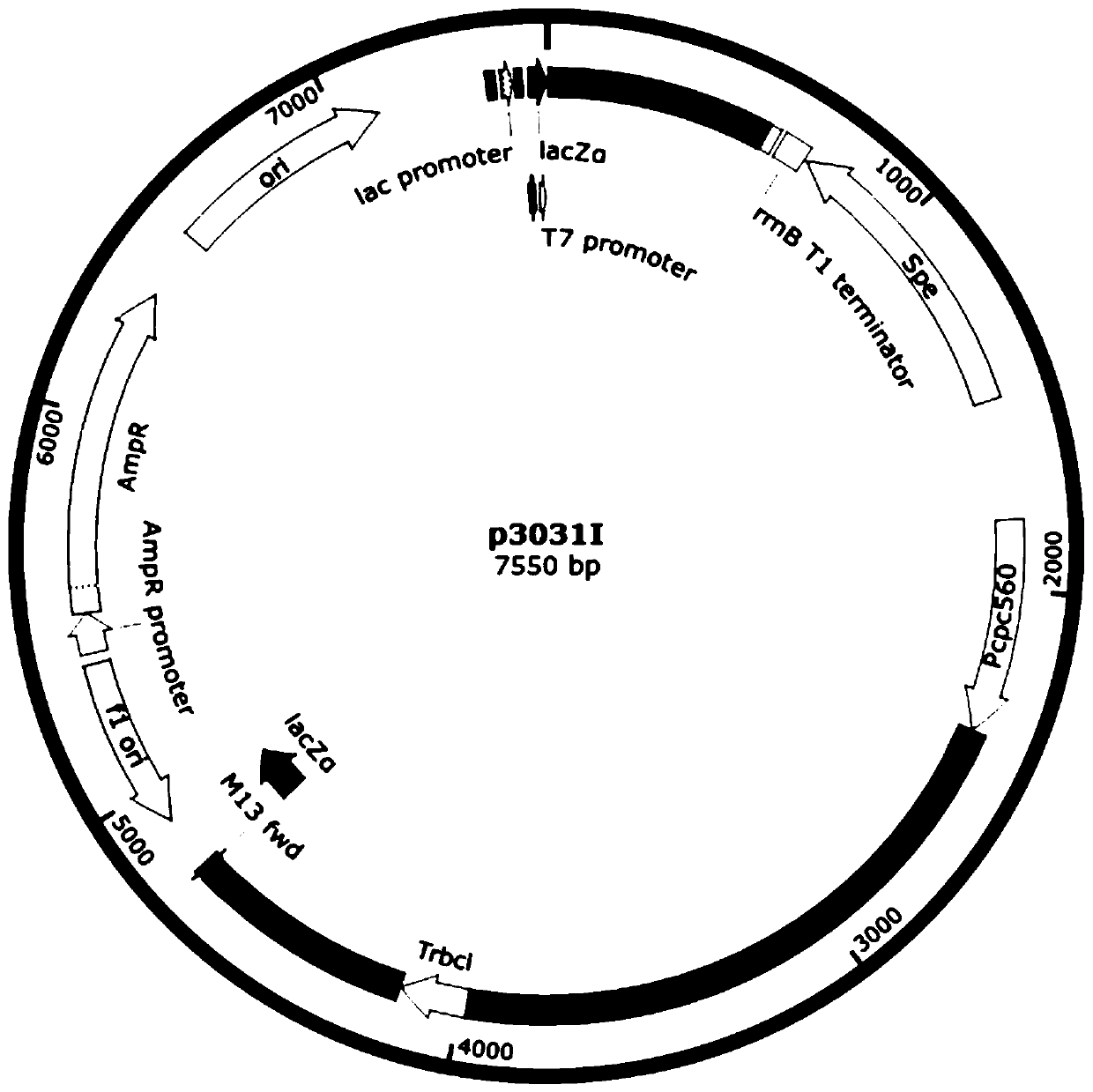
[0039] (2) Using the nucleotide sequences shown in SEQ ID NO.12 and SEQ ID NO.13 in the sequence listing as upstream and downstream primers, using the plasmid pCP3031 containing the cpc560 promoter and rbcL terminator as a template, amplified to obtain the third Backbone fragment, the third skeleton fragment and the INO1 gene fragment obtained in step (1) are bluntly connected to construct the recombinant expression vector p3031I, with the nucleotides shown in SEQ ID NO.14 and SEQ ID NO.9 in the sequence listing Sequences were verified for upstream and downstream primers.
[0040] (3) Use the nucle...
Embodiment 2
[0048] Construction of strain WT-INO1SSZP.
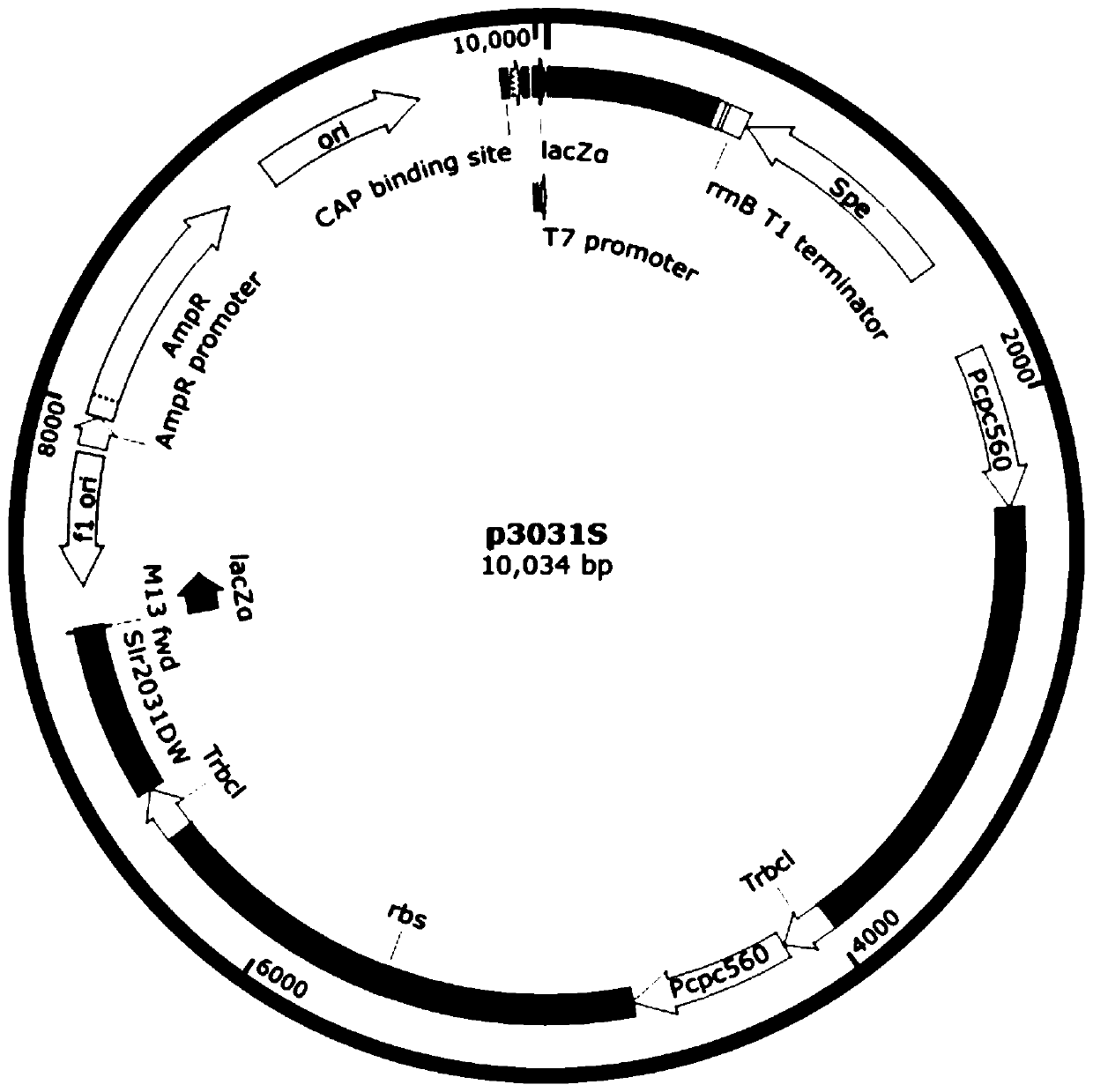
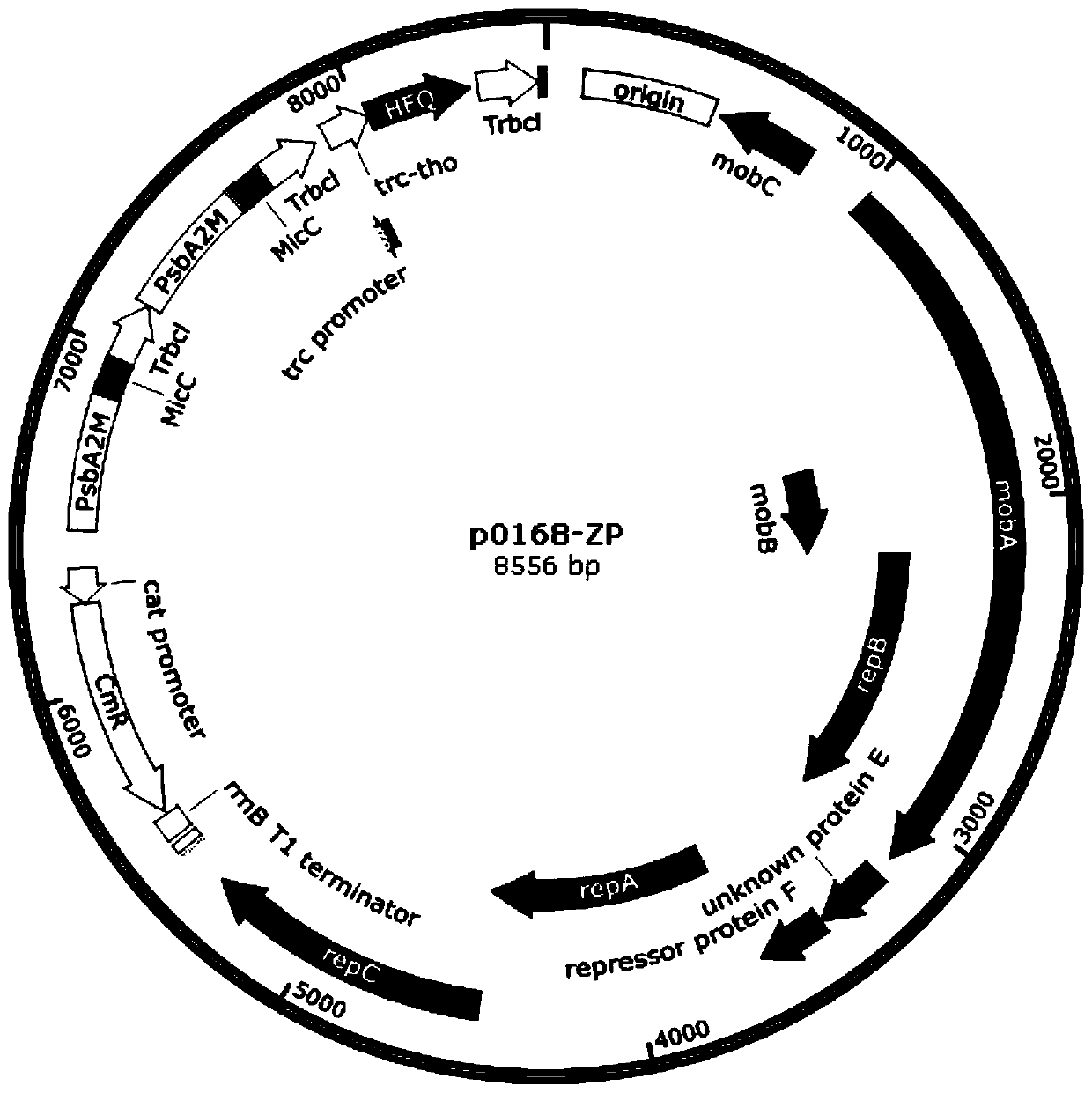
[0049] (1) Using the nucleotide sequences shown in SEQ ID NO.23 and SEQ ID NO.24 in the sequence listing as upstream and downstream primers, and using the plasmid pBA3031-HM(the*) as a template, perform PCR reverse amplification to obtain The first skeleton fragment, the first skeleton fragment is self-ligated to obtain the recombinant expression vector P3;
[0050] Using the nucleotide sequences shown in SEQ ID NO.25 and SEQ ID NO.24 as upstream and downstream primers, and using the plasmid pBA3031-HM(the*) as a template, PCR reverse amplification was performed to obtain the second backbone fragment, the The two backbone fragments were self-ligated to obtain the recombinant expression vector P4.
[0051] (2) Use the nucleotide sequences shown in SEQ ID NO.8 and SEQ ID NO.9 in the sequence table as upstream and downstream primers, and use the recombinant expression vector P3 as a template to perform PCR amplification to obtain Ppsb...
Embodiment 3
[0058] Optimization of Glucose Supplement in Culture Conditions of Genetically Engineered Bacteria WT-INO1SSZP
[0059] Inoculate the genetically engineered bacteria WT-INO1SSZP in a 100mL shake flask (initial OD of bacterial solution 730 nm=0.1, the medium volume after inoculation is 25mL), at 150rpm, 30℃, 50μmol photons m -2 the s -1 Under the conditions, 5mM glucose was added to the medium every 24h. After culturing the WT-INO1SSZP strain for three days, all the cells were centrifuged at 4°C and 3000×g for 10 min to collect the cells, and the collected cells were resuspended in fresh BG-11 liquid medium with a final concentration of 2 mM theophylline. The resuspended cell culture was continued for 5 days, and then the inositol production was measured. After testing, it was found that WT-INO1SSZP was optimized by adding glucose, and the production of inositol reached 21.74mg L after 8 days of culture -1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com