Photoelectric chemical biosensor as well as preparation method and application of photoelectric chemical biosensor
A biosensor, photoelectrochemical technology, applied in the field of sensors, can solve the problems of limited photoelectric signal and sensitivity of photoelectric sensors, and achieve the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
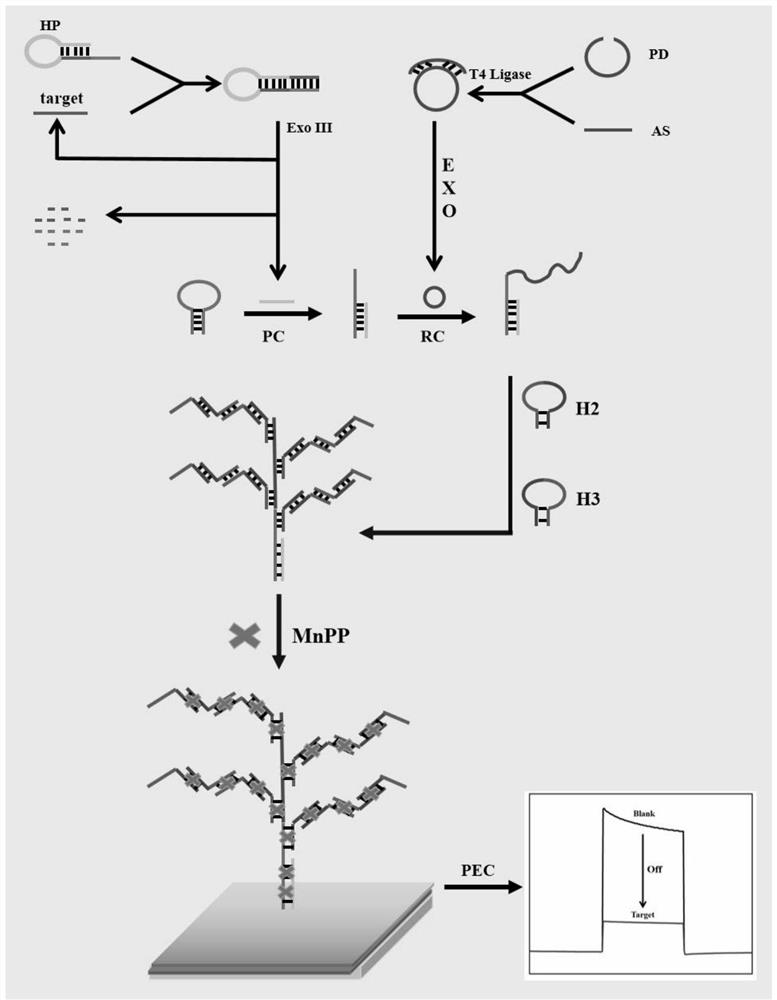
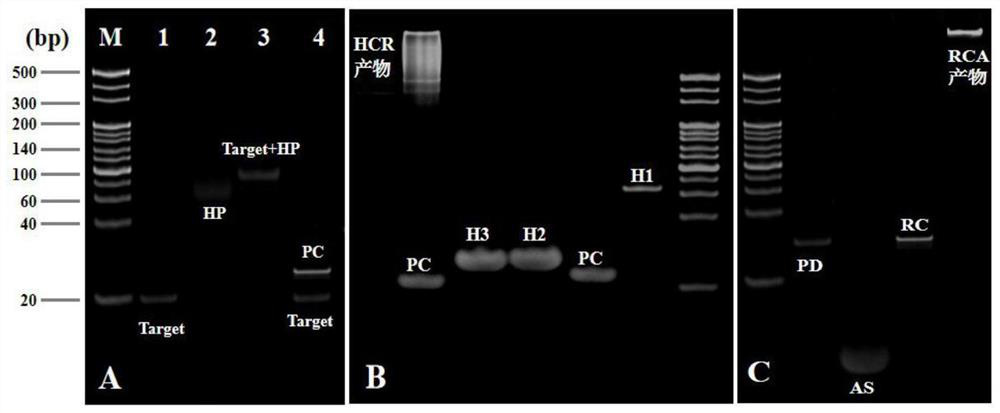
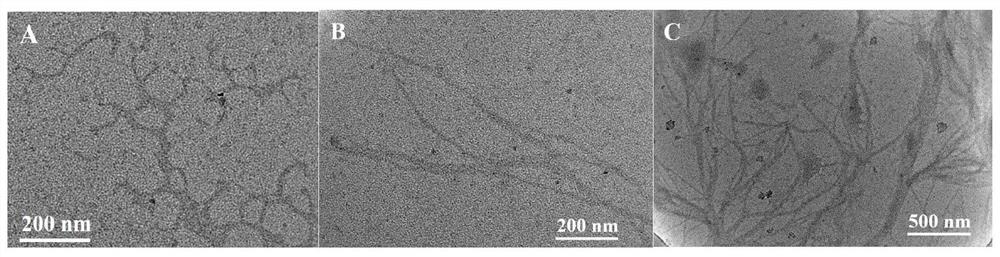
[0055] Mix 1 μL of padlock DNA with a concentration of 100 μM, 3 μL of assistantDNA with a concentration of 100 μM, 4 μL of secondary water and 3 μL of a buffer solution of 10×T4 DNA ligase, and anneal at 95°C for 5 min, then naturally cool to room temperature, and then add 120 U of T4DNA ligase was added to the mixed solution and connected at 37°C for 1h, then the mixed solution was reacted at 65°C for 10min to inactivate T4DNA ligase, after cooling to room temperature, add 4μL exonuclease buffer solution, 20U exonuclease I and 100U exonuclease III, cutting at 37°C for 2h, then reacting the mixed solution at 80°C for 15min to inactivate exonuclease I and exonuclease III, after cooling to room temperature, the DNA rolling circle template can be obtained , and store the obtained DNA rolling circle template at 4°C for future use.
[0056] Anneal 18 μL of HP with a concentration of 1 μM at 95°C for 5 minutes, then cool naturally to room temperature to obtain the hairpin structure...
Embodiment 2
[0059] Mix 1 μL of padlock DNA with a concentration of 100 μM, 3 μL of assistantDNA with a concentration of 100 μM, 4 μL of secondary water and 3 μL of a buffer solution of 10×T4 DNA ligase, and anneal at 97°C for 4 min, then naturally cool to room temperature, and then add 130 U of T4DNA ligase was added to the mixed solution and connected at 37°C for 2h, then the mixed solution was reacted at 68°C for 8min to inactivate T4DNA ligase, after cooling to room temperature, add 4μL exonuclease buffer solution, 30U exonuclease I and 110U exonuclease III, cutting at 37°C for 2h, then reacting the mixed solution at 83°C for 12min to inactivate exonuclease I and exonuclease III, after cooling to room temperature, the DNA rolling circle template can be obtained , and store the obtained DNA rolling circle template at 6°C for future use.
[0060] Anneal 18 μL of HP with a concentration of 1 μM at 97°C for 4 minutes, then cool naturally to room temperature to obtain the hairpin structure,...
Embodiment 3
[0063] Mix 1 μL of padlock DNA with a concentration of 100 μM, 3 μL of assistantDNA with a concentration of 100 μM, 4 μL of secondary water and 3 μL of a buffer solution of 10×T4 DNA ligase, and anneal at 93 °C for 6 min, then naturally cool to room temperature, and then add 1110 U of T4DNA ligase was added to the mixed solution and connected at 35°C for 1h, then the mixed solution was reacted at 62°C for 12min to inactivate T4DNA ligase, after cooling to room temperature, add 4μL exonuclease buffer solution, 10U exonuclease I and 90U exonuclease III, cutting at 35°C for 1h, then reacting the mixed solution at 77°C for 18min to inactivate exonuclease I and exonuclease III, after cooling to room temperature, the DNA rolling circle template can be obtained , and store the obtained DNA rolling circle template at 2°C for future use.
[0064] Anneal 18 μL of HP with a concentration of 1 μM at 93°C for 6 minutes, then cool naturally to room temperature to obtain the hairpin structur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com