Genetically engineered bacteriophage
A bacteriophage and gene technology, applied in the direction of bacteriophage, virus/bacteriophage, medical raw materials derived from virus/bacteriophage, etc., can solve the problem of bacteriophage DNA damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0164] Example 1-Phage isolation
[0165] Collect and purify samples from sewers and waste, environmental soil and animal feces for the isolation of bacteriophages. The purified sample is then screened for the presence of phages against specific bacteria. According to Bonilla et al. (2016) and Bourdin et al. (2014), the scheme and method of separating bacteriophages from water samples was adopted; according to Silankorva (2018), Pausz et al. Solutions and methods.
[0166] Rehydrate the solid sample with sterile water for at least 1 hour to allow the phage to spread. The sample is then centrifuged to remove solid matter and large particles, and the supernatant is collected. The centrifuged environmental samples and water samples are then further processed and purified using a filter (0.2 μM) to remove bacteria and smaller harmful particles. The filtered sample can be further concentrated using a filter tube, or stored at 4°C for future use.
[0167] The filtered samples were the...
example 2
[0169] Example 2-Platform Development
[0170] Using environmental samples EV31 / PP8, after phage isolation, the genomic material was purified using PureLink viral DNA / RNA extraction kit. Amplify the full-length genome (EV31 / Full / F / pYESIL and EV31 / Full / R / pYESIL, see sequence EV31) to have 30bp homology with the pYESIL sapphire vector. Phusion high-fidelity DNA polymerase was used for PCR amplification (modification using touchdown technology, the primers were annealed from 69°C, and each cycle decreased by 0.5°C). The PCR products were separated on an agarose gel, and the bands were cut, extracted and assembled. The resulting construct EV31pYES (unmodified) can be used for genetic modification of EV31 and the determination of mutation function in a phage rescue-based system.
example 3
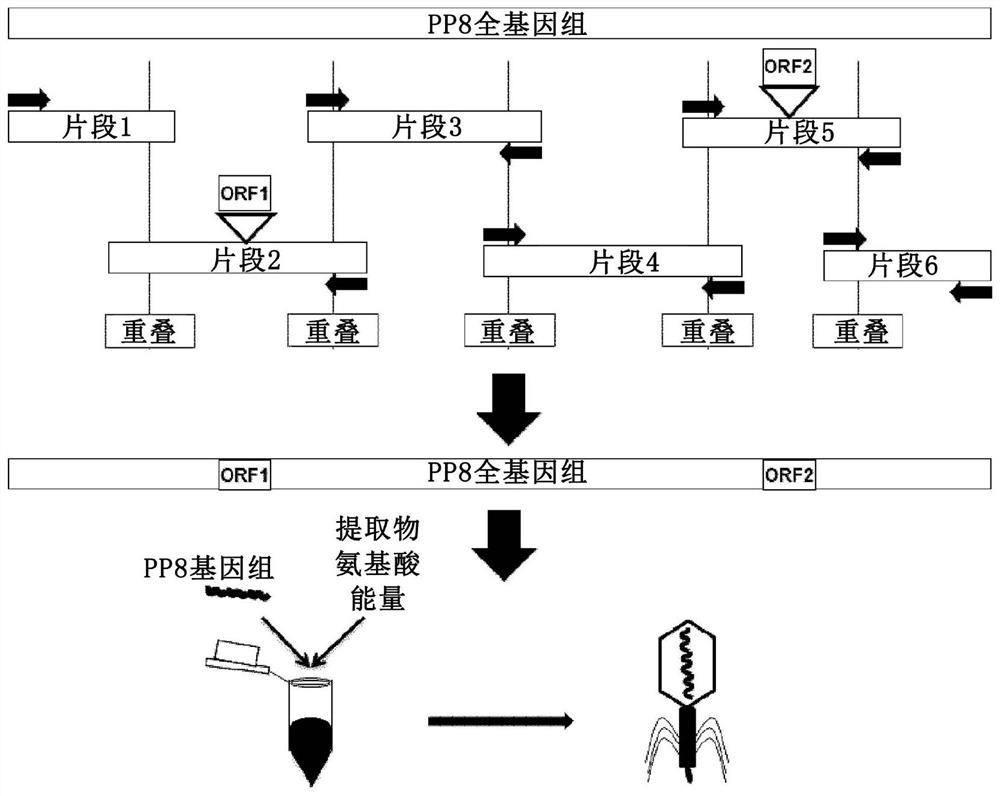
[0171] Example 3-Full-length genome assembly
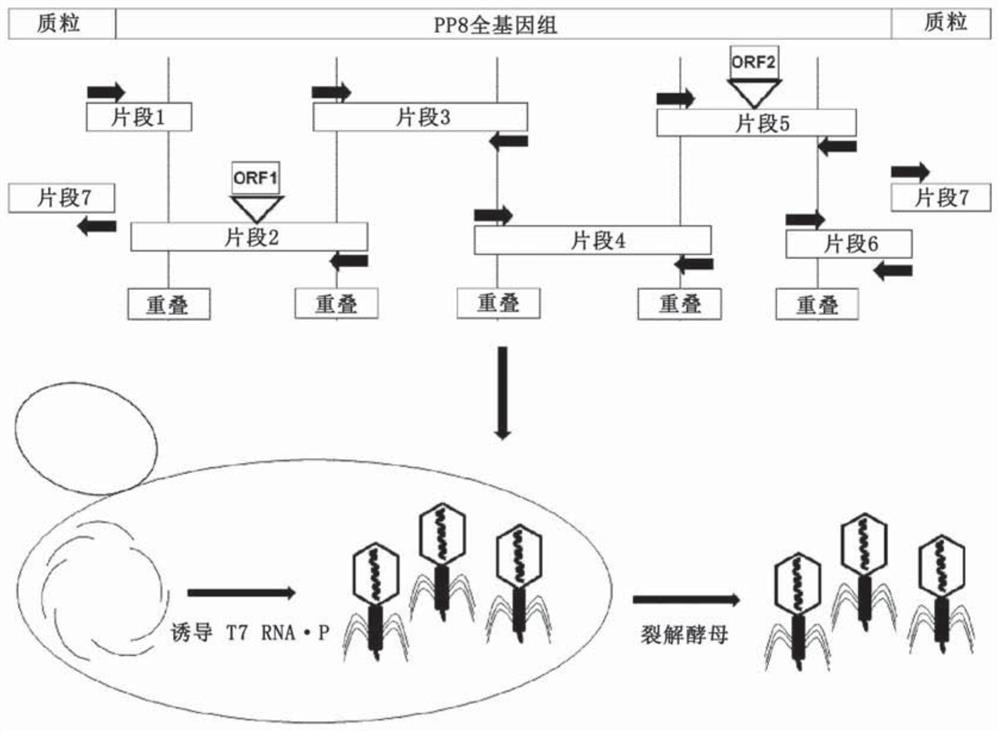
[0172] In short, the following provides a method for genetic manipulation of yeast (Kluyveromyces lactis and Pichia pastoris) cells to include T7 DNA (deoxyribonucleic acid) dependent from E.coli phage T7 The RNA (ribonucleic acid) polymerase is transcribed, and then the phage is expressed in yeast. A method for genetic manipulation of yeast (Kluyveromyces lactis and Pichia pastoris) cells is also provided, so that it includes bacteria (E. coli) and RNA (ribonucleic acid) polymerase (P ) And then express the phage in yeast.
[0173] See image 3 , Divide the genomic complement into fragments that overlap with adjacent fragments obtained by PCR amplification. Insert foreign genes into their respective fragments. The fragments were combined into a full-length genome by homologous recombination, and the yeast-based plasmid (as an additional PCR fragment) was combined with the T7 promoter in the yeast strain Pichia pastoris. The stable...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com