Visual distinguishing method for single-cell DNA epigenetic modification space positioning and adjacent distribution
A technology of epigenetic modification and spatial positioning, which is applied in the field of visual distinction between the spatial positioning and adjacent distribution of single-cell DNA epigenetic modification, and can solve the problems of identification interference and inability to obtain information on the spatial adjacent distribution of DNA epigenetic marks. , to achieve the effect of avoiding interference and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
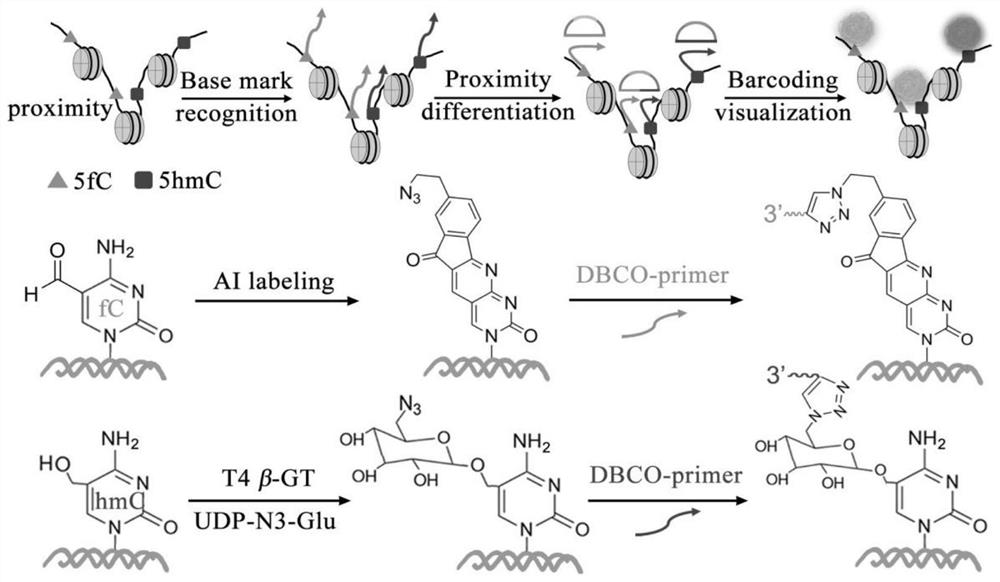
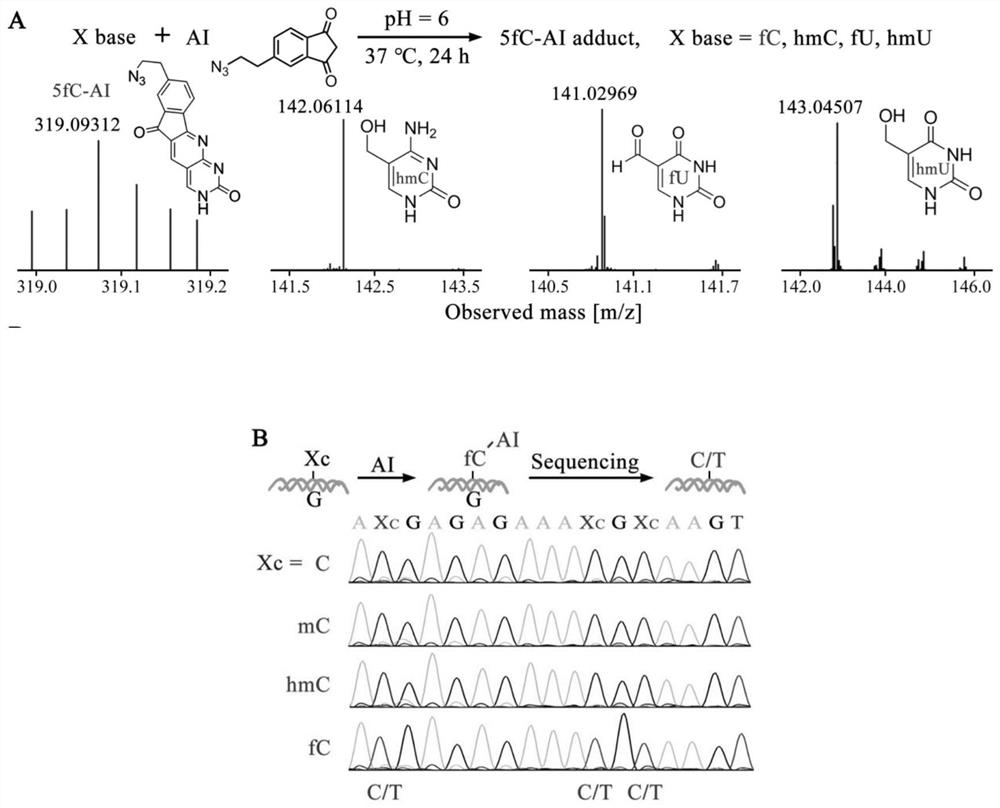
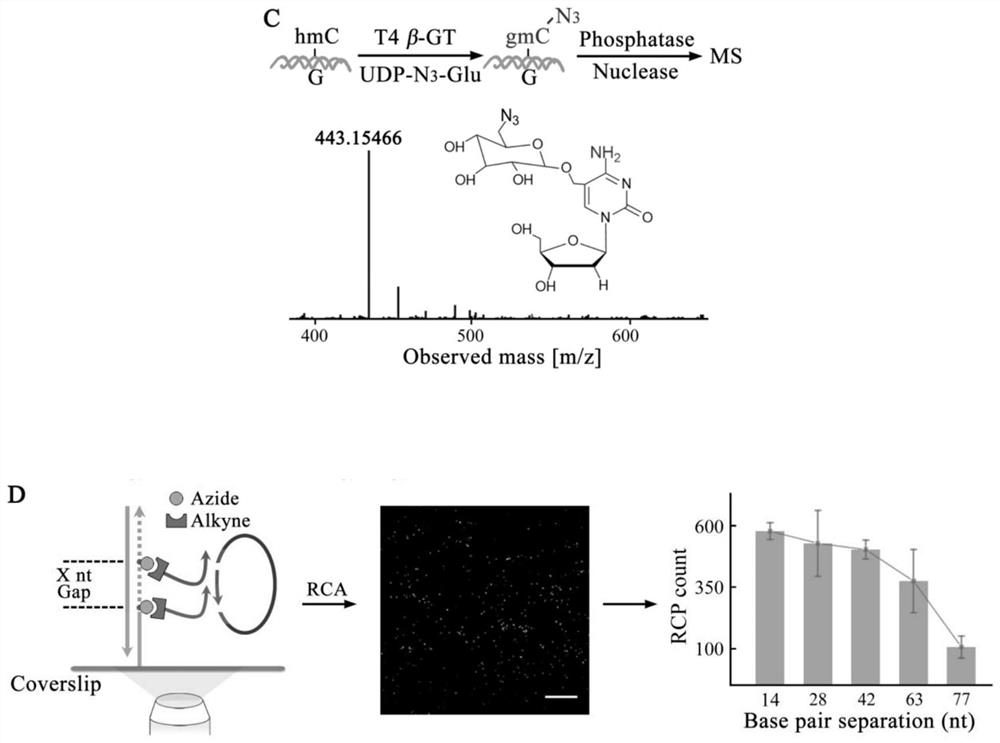
[0042] Example 1: Combination figure 1 , Visualization of the spatial location and neighboring distribution of DNA epigenetic modifications in MCF-10A cells. The cells were fixed on a cover glass with a micro reaction chamber using paraformaldehyde.
[0043] N of 5-fC by azide derivative probe AI 3 Chemical labeling of functional groups, the use of β-GT for 5-hmC N 3 Enzyme-mediated labeling of functional groups. 20 μL of 1×MES buffer containing 10 mM AI and 10% dimethyl sulfoxide was added to the reaction chamber and incubated at 37° C. for 24 h. Then, using click chemistry reaction, 1×PBS buffer containing 100 nM DBCO modified primer probe P1-fC was added to the reaction chamber, and the reaction was carried out at 37° C. for 1 h. Then add 20μL of UDP-N containing 1×NEBuffer 4, 50μM to the reaction chamber 3 -Glu and 5U T4β-GT culture solution, incubated at 37°C for 2h. Finally, 100nM P2-hmC probe was added to the reaction chamber and reacted at 37°C for 1h. Remove the react...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com