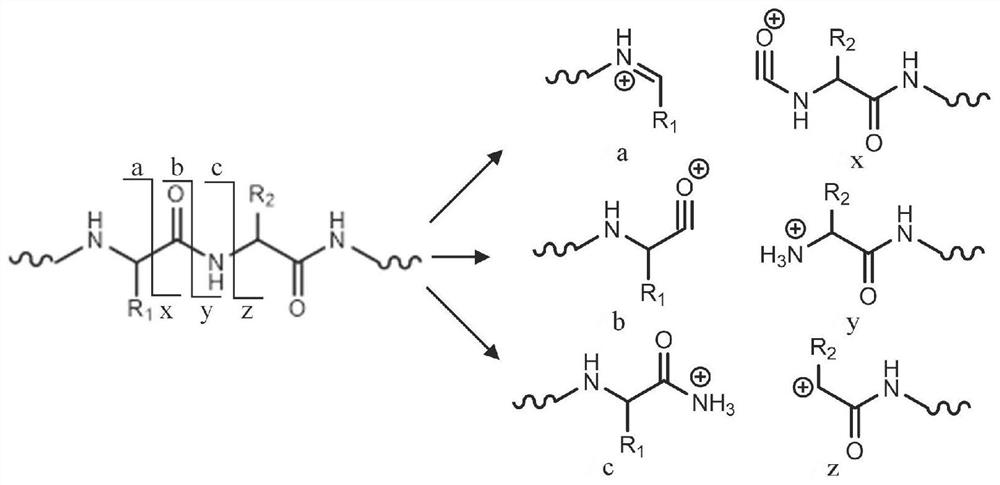
Sequence analysis method and identification method of insulin or analogue thereof
A technology for sequence analysis and insulin, applied in the field of sequence analysis method and identification of insulin or its analogs, can solve the problems of artificial modification of enzymatic hydrolysis reaction, long enzymatic hydrolysis reaction time, occurrence of interference peaks, etc., and achieves rapid reduction reaction and identification. Fast, accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The sequence analysis of embodiment 1 insulin aspart
[0070] Insulin aspart injection was diluted to 1 mg / mL with 10 mmol / L hydrochloric acid solution, 100 μL was taken into a centrifuge tube, and final concentrations of 6 mol / L guanidine hydrochloride and 50 mmol / L TCEP were added, and after incubation at 45°C for 40 min, the aspart The disulfide bond of insulin is reduced and opened, and the two peptide chains are separated. The reduced product was analyzed by ultra-high performance liquid chromatography-high resolution tandem mass spectrometry, and the injection volume was 2 μL. The instrumental analysis conditions are as described above. The preferred post-mass fragmentation mode of the insulin aspart A chain is HCD, and the preferred post-collision energy is 20; the preferred post-mass fragmentation mode of the insulin aspart B chain is HCD, and the preferred post-collision energy is 25.
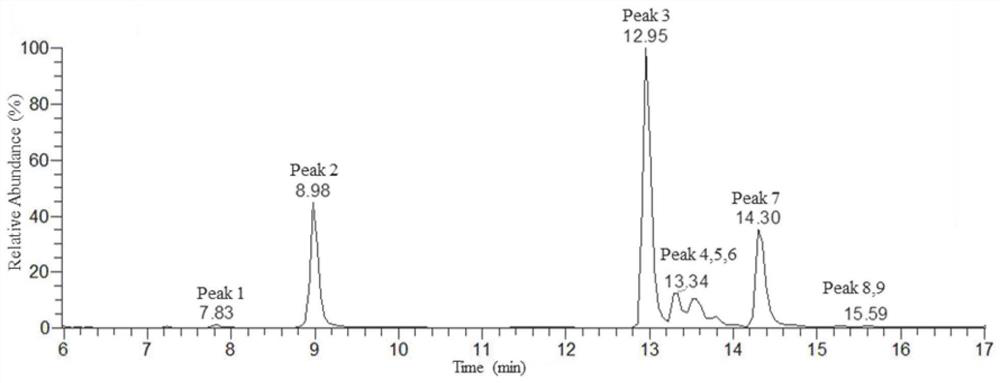
[0071] The detection results of liquid chromatography-mass spectrometry a...
Embodiment 2
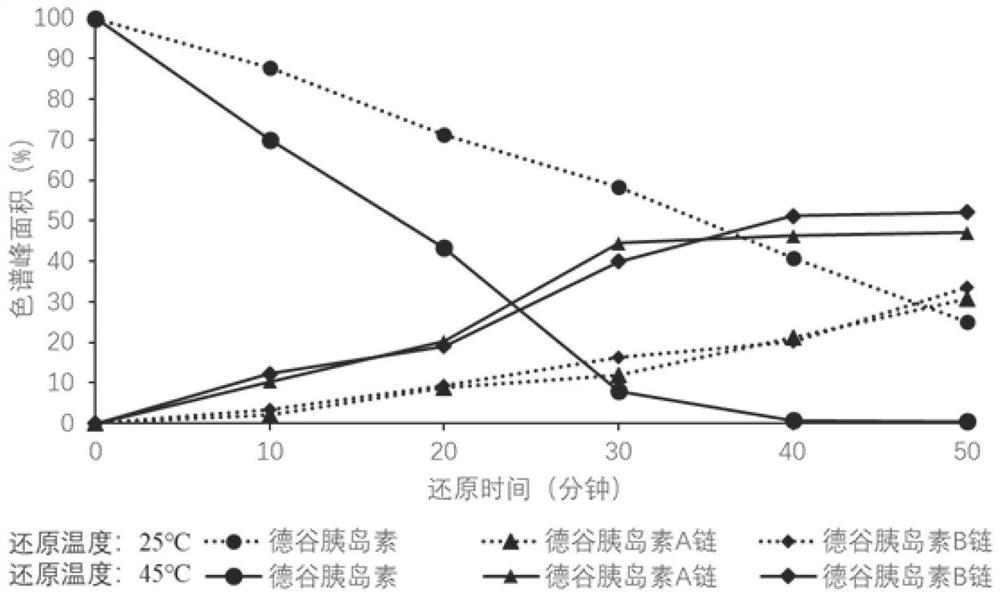
[0087] Example 2 Sequence Analysis of Insulin Degludec
[0088] Dilute insulin degludec injection with 10mmol / L hydrochloric acid solution to 1mg / mL, take 100μL into a centrifuge tube, add 6mol / L guanidine hydrochloride and 50mmol / L TCEP at final concentrations respectively, incubate at 45°C for 40min, and make degludec The disulfide bond of insulin is reduced and opened, and the two peptide chains are separated. The reduced product was analyzed by ultra-high performance liquid chromatography-high resolution tandem mass spectrometry, and the injection volume was 2 μL. The instrumental analysis conditions are as described above. The preferred post-mass fragmentation mode of the insulin degludec A chain is HCD, and the preferred post-collision energy is 20; the preferred post-mass fragmentation mode of the insulin degludec B chain is HCD, and the preferred post-collision energy is 25.
[0089] The detection results of liquid chromatography-mass spectrometry are as follows: F...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com