A method for high-throughput screening of DNA damage response inhibitors
A technology of DNA damage and reaction inhibitors, applied in neural learning methods, drugs or prescriptions, image enhancement, etc., can solve the problems of laborious and error-prone marking Foci, achieve high accuracy and reduce time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] The technical scheme of the present invention will be described in further detail below in conjunction with the accompanying tables and examples. The following examples are implemented on the premise of the technical solutions of the present invention, and provide detailed embodiments and processes, but the protection scope of the present invention is not limited to the following examples.
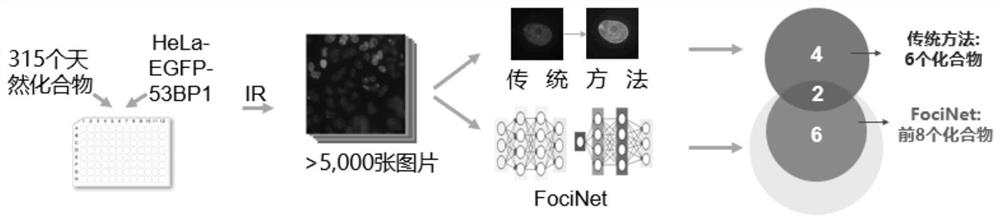
[0030] like figure 1 As shown, this example provides a method for high-throughput screening of DNA damage response inhibitors.
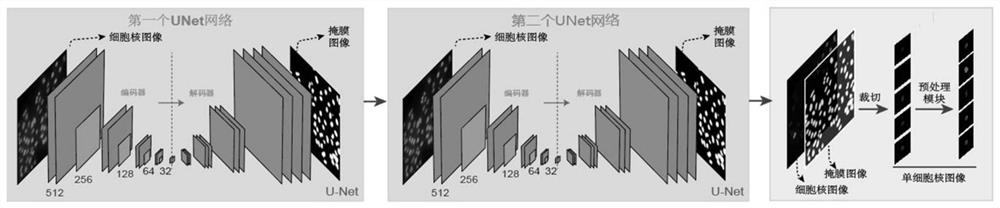
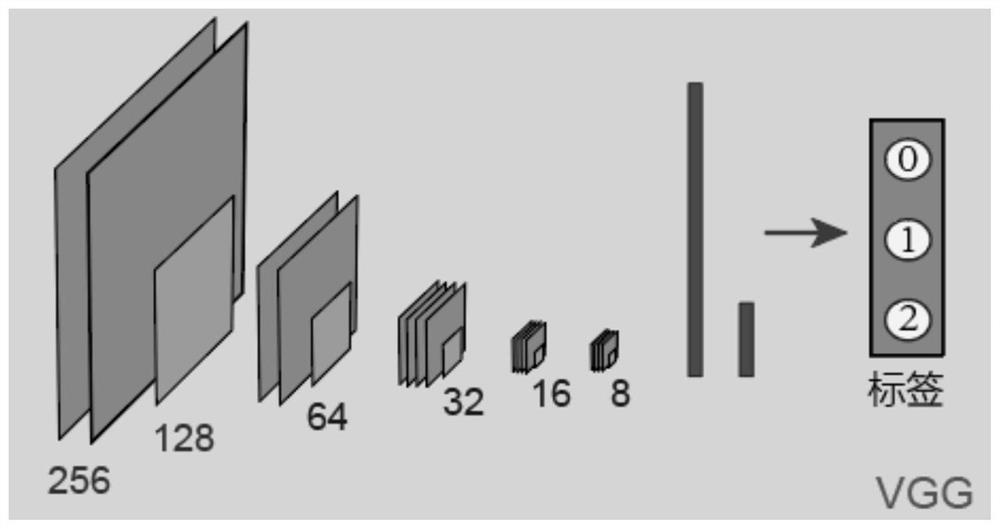
[0031] S1. Train the cell nucleus segmentation network model based on the U-Net network.
[0032] The cell nucleus segmentation network model is used to segment the image data captured by the high-content imaging device, and after obtaining a mask image corresponding to the image data, the mask image is used to crop the original image to obtain a single cell nucleus image.
[0033] S11. Construct a first U-Net network and a second U-Net network.
[0034...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com