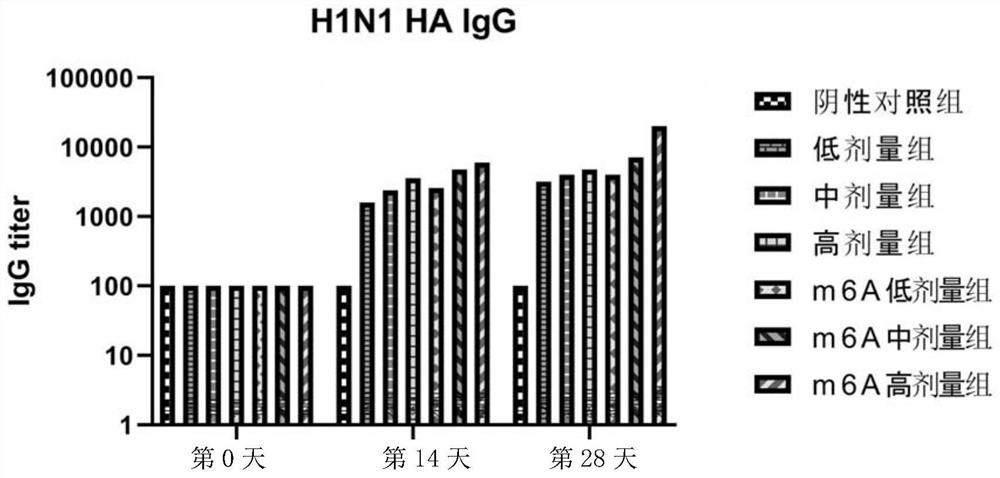
Circular RNA vaccine for influenza viruses
A circular and vaccine technology, applied in the field of circular RNA vaccines, can solve the problems of the advent of mRNA vaccines and poor stability of mRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1, construction recombinant plasmid
[0042] Through the analysis of a large number of strain sequences, the H1N1-WSN-HA antigen was truncated and modified, and multiple point mutations were introduced based on virus variation to integrate H1N1-M2e antigen, H3N2-M2e antigen, H5-M2e (H5N1, H5N6) antigen, H7N9- For the M2e antigen and the H9N2-M2e antigen, the segments are connected by GGGGS, and the protein shown in sequence 1 of the sequence table is designed. The interleukin-2 signal peptide is added to the N-terminal of the protein shown in sequence 1 in the sequence listing, and the Flag tag is added to the C-terminal to design the protein shown in sequence 2 in the sequence listing.
[0043] Through codon optimization, the DNA molecule shown in sequence 3 of the sequence listing is obtained, which encodes the protein shown in sequence 2 of the sequence listing. The IRES sequence is added downstream of the DNA molecule shown in sequence 3 in the sequence ...
Embodiment 2
[0046] Embodiment 2, preparation circularized RNA
[0047] 1. Take the recombinant plasmid IAV-circmRNA-1.0-pBluescript II KS(+), digest it with restriction endonuclease XhoI, and recover the linearized plasmid.
[0048] 2. Transcribing RNA
[0049] Reaction system (20 μl): 1 μg linearized plasmid, 2 μl NTP mixture, 2 μl 10× reaction buffer and T7 transcriptase, the balance is DEPC water. The active ingredients provided by the NTP mix are ATP, CTP, GTP and UTP. In the reaction system, the concentrations of ATP, CTP, GTP and UTP are all 100mM, and the content of T7 transcriptase is 100U. NTP mixture: Beijing Shengke Boyuan Biotechnology Co., Ltd., catalog number 70906. 10× reaction buffer: Shanghai Yuanmu Biotechnology Co., Ltd., catalog number YM-MY162J. T7 transcriptase: Beijing Biolab Biotechnology Co., Ltd., catalog number JN0010.
[0050] Reaction conditions: 37°C water bath, 16h.
[0051] 3. After completing step 2, use QIAGEN's RNA purification kit for purification ...
Embodiment 3
[0062] Example 3. Preparation of HA2-M2e protein using HA2-M2e circ mRNA
[0063] 1. Take the recombinant plasmid IAV-circmRNA-1.0-pBluescript II KS(+), digest it with restriction endonuclease XhoI, and recover the linearized plasmid.
[0064] 2. Transcribed and purified RNA
[0065] Take step 1 to get the linearized plasmid, use HiScribe TM T7 High Yield RNA Synthesis Kit (New England Biolabs (UK) Ltd) was used for in vitro transcription, and the specific operation steps were carried out according to the instructions.
[0066] 3. Take the product of step 2, and use RNeasy Mini Kit (QIAGEN) to purify to obtain purified RNA.
[0067] 4. Cyclization identification
[0068] Method is the same as step 6 of embodiment 2.
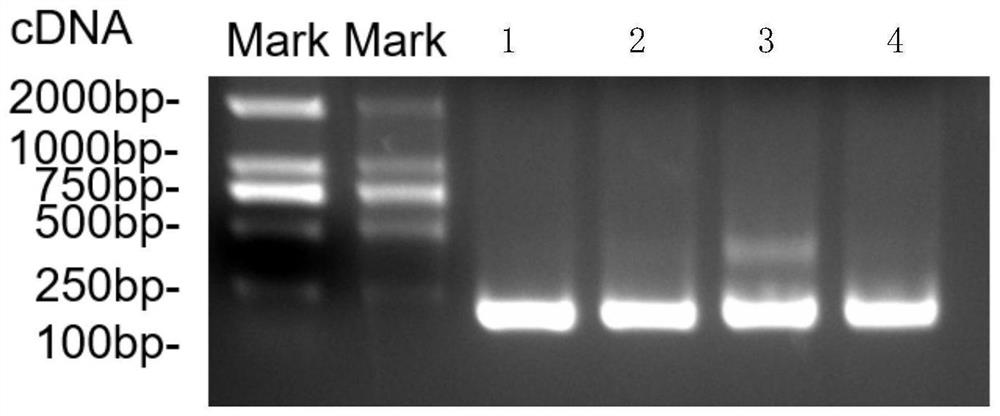
[0069] Electropherogram see figure 1 Lane 2 of , shows a target band of about 250bp, indicating that circularized RNA was obtained.
[0070] Therefore, the purified RNA obtained in step 3 was named HA2-M2e circ mRNA.
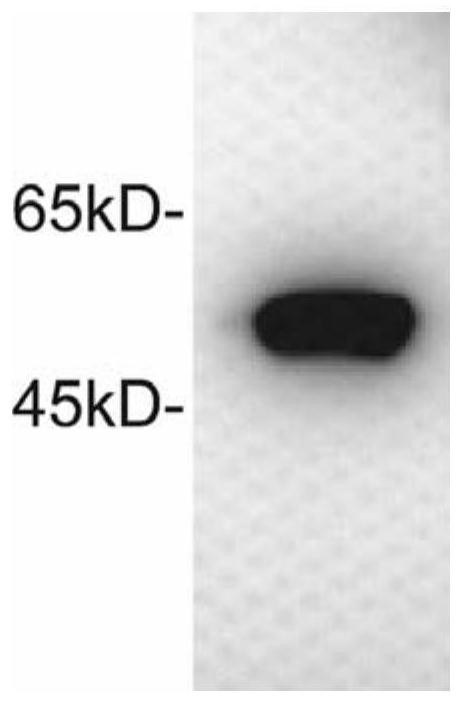
[0071] 5. In vitro expression identif...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com