Nucleic acid aptamer delivery vector and preparation method and application thereof
A nucleic acid aptamer and delivery carrier technology, applied in the field of molecular biology, can solve the problems of complex separation methods, high cost, and low yield of natural exosomes, and achieve high biocompatibility, easy operation, and low equipment prices Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
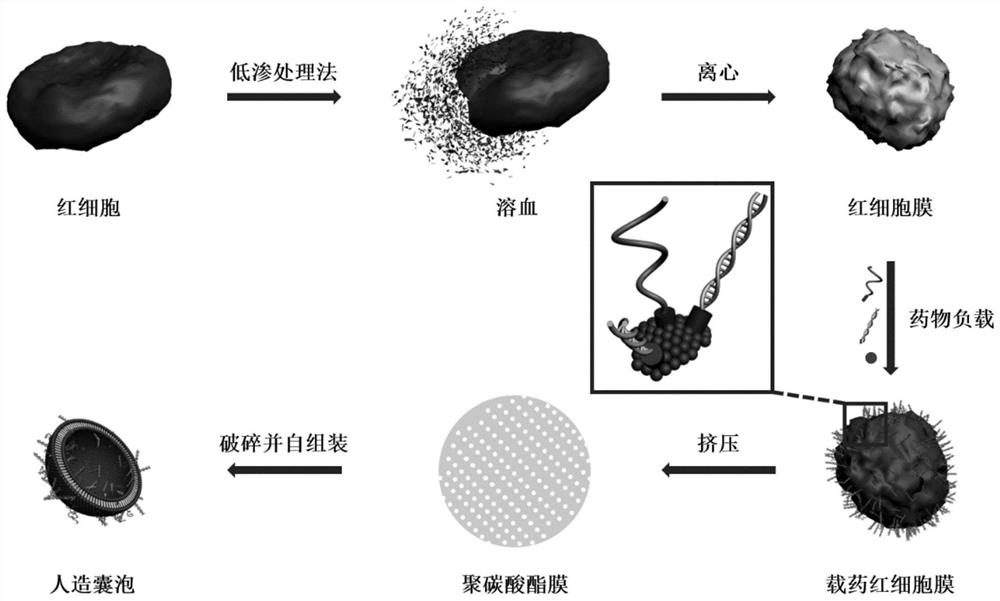
[0071] The embodiment of the present invention provides a small nucleic acid delivery carrier based on aptamer targeting, its preparation method and its drug loading method, including:
[0072] Using hypotonic treatment to prepare red blood cells into red blood cell membranes;
[0073] Preparation of red blood cell membranes into nanovesicles by extrusion through the membrane;
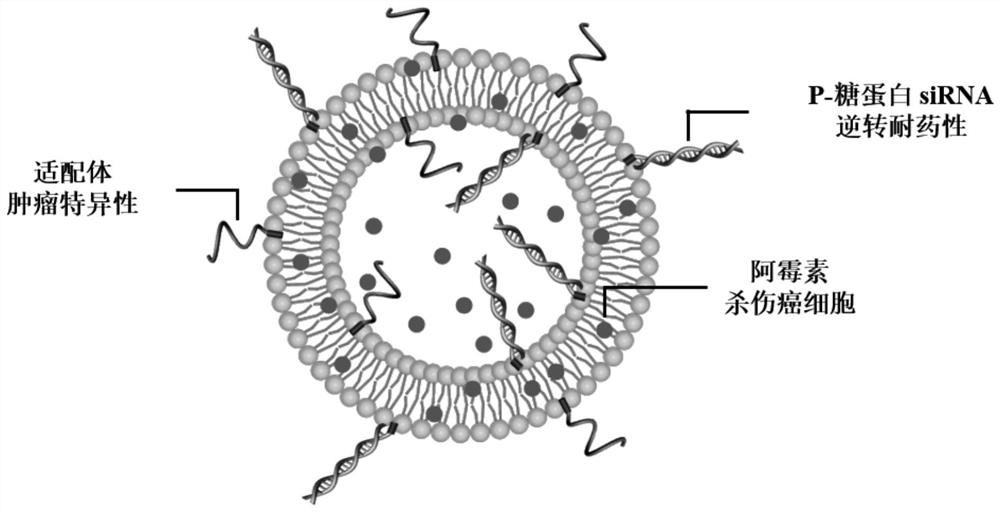
[0074] The cholesterol-modified aptamer, the cholesterol-modified small nucleic acid and the chemotherapeutic drug doxorubicin are loaded onto the membrane of nanovesicles by incubation to prepare targeted drug-loaded nanovesicles.
[0075] Wherein, the red blood cells are placed in PBS solution and incubated with shaking for 1-3 hours, and centrifuged at 12000-14000 rpm for 10-20 minutes to collect red blood cell membranes. The erythrocyte membrane is ultrasonically broken, and extruded through the membrane for 10 to 30 times to obtain nano-sized vesicles.
[0076] Among them, the ultrasonic breaker...
Embodiment 1
[0119] Step 1: Add red blood cells to 1×PBS, centrifuge at 800g at 4°C for 5 minutes, collect red blood cells, and repeat twice. Then red blood cells were added to 0.25×PBS, incubated on ice for 1 hour with shaking, 14000 rpm, and centrifuged at 4°C for 10 minutes to collect red blood cell membranes. Then add 0.25×PBS, shake and incubate on ice for 1h, centrifuge at 14000rpm, 4°C for 10min, and collect the red blood cell membrane.
[0120] Among them, the volume ratio of red blood cells and 0.25×PBS is 1:9.
[0121] Step 2: Place the collected erythrocyte membrane in an ultrasonic breaker for ultrasonic crushing, the conditions are 60W power, 25KHz frequency, horn model Φ2, ultrasonication for 1min. Then use an Avanti miniextruder liposome extruder equipped with a polycarbonate membrane with a pore size of 200 nm, and extrude the sonicated red blood cell membrane 30 times to obtain nanovesicles.
[0122] Steps 1 to 2 above can be performed by figure 1 express.
[0123] The...
Embodiment 2
[0125] Step 1: Add red blood cells to 1×PBS, centrifuge at 800g at 4°C for 5 minutes, collect red blood cells, and repeat twice. Then red blood cells were added to 0.25×PBS, incubated on ice for 3 hours with shaking, 14000 rpm, and centrifuged at 4°C for 10 minutes to collect red blood cell membranes. Then add 0.25×PBS, shake and incubate on ice for 1h, centrifuge at 12000rpm, 4°C for 20min, and collect the red blood cell membrane.
[0126] Among them, the volume ratio of red blood cells and 0.25×PBS is 1:9.
[0127] Step 2: Place the collected erythrocyte membrane in an ultrasonic breaker for ultrasonic crushing, the condition is 20W power, 20KHz frequency, horn model Φ2, ultrasonic 5min. Then use an Avanti miniextruder liposome extruder equipped with a polycarbonate membrane with a pore size of 100 nm, and extrude the sonicated red blood cell membrane 10 times to obtain nanovesicles.
[0128] Steps 1 to 2 above can be performed by figure 1 express.
[0129] The nanovesic...
PUM
| Property | Measurement | Unit |
|---|---|---|
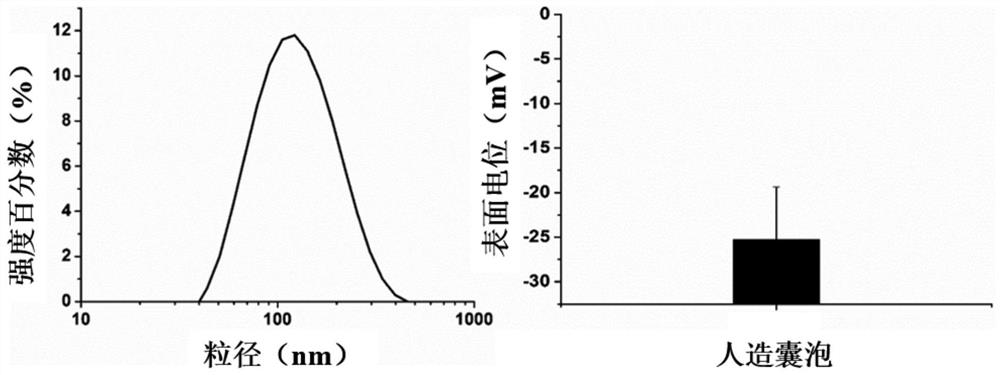
| Particle size | aaaaa | aaaaa |
| Concentration | aaaaa | aaaaa |
| Aperture | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com