Blocking or weakening the expression of rice circRNA coding site to improve the method of rice seedling growth traits
A technology for growth traits and coding bits, which is applied in the field of plant biology to achieve the effects of easy operation, good quality improvement and concise steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
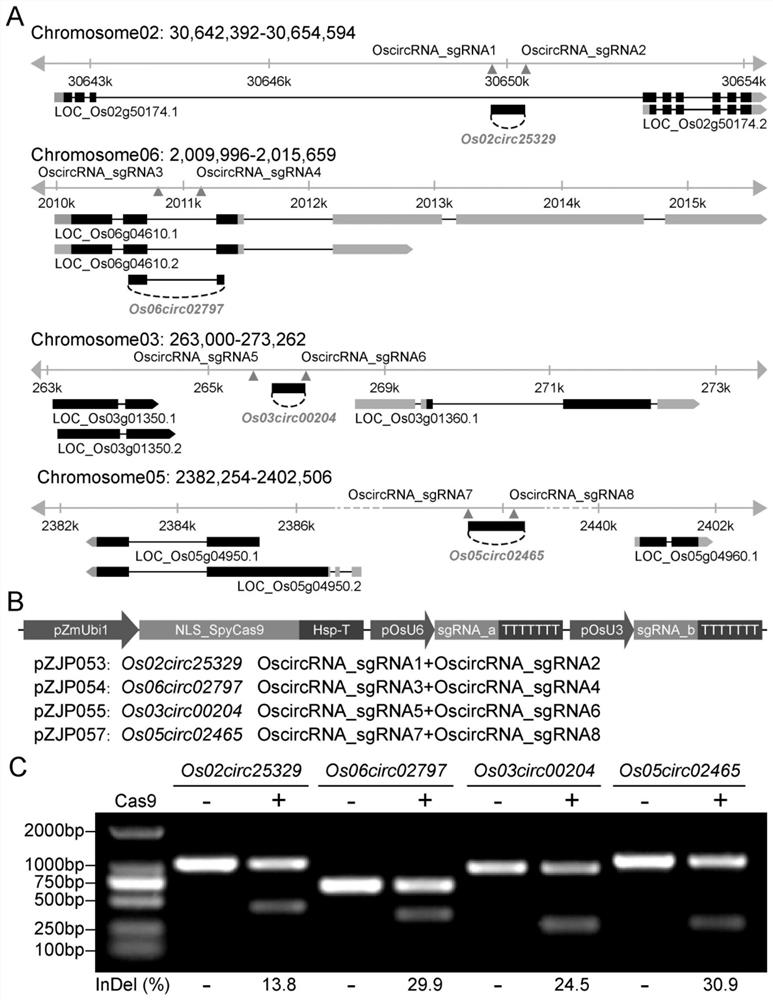
[0099] Example 1 Construction of rice Os06circ02797 knockout editing vector pZJP054
[0100] (1) sgRNA design
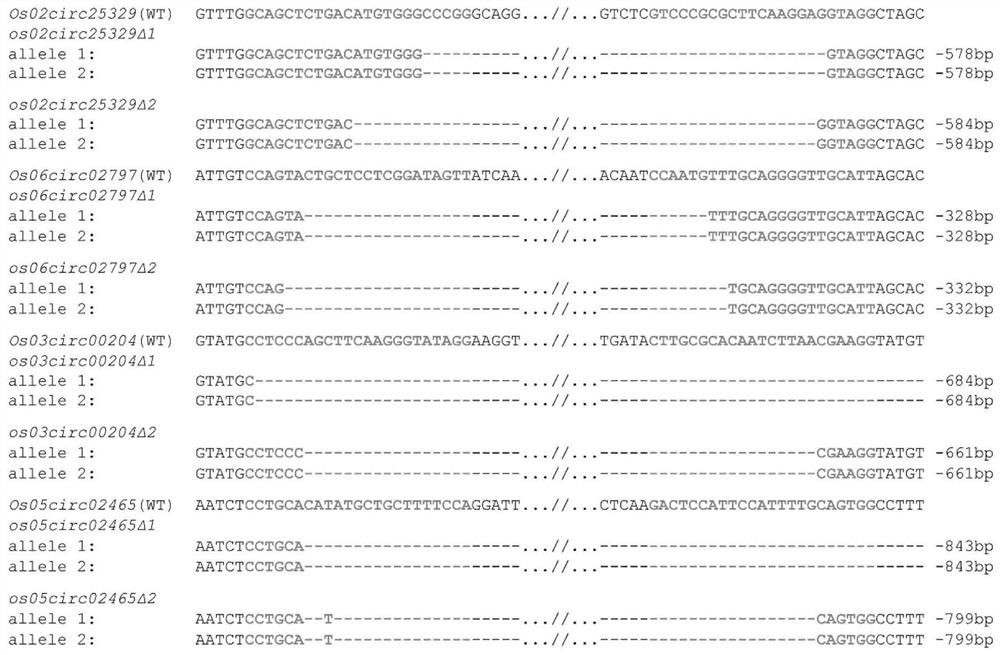
[0101] From the database website NCBI (https: / / www.ncbi.nlm.nih.gov / ), search and download the rice circRNA coding site Os06circ02797 sequence, and then according to the recognition and cutting rules of the target site by the CRISPR-Cas9 system, Design the sgRNA; then use the CRISPR-P website online (http: / / crispr.hzau.edu.cn / CRISPR2 / ) to predict the mismatch rate and off-target sites of the sgRNA and select the optimal sgRNA3 (SEQ ID No.1 ) and sgRNA4 (SEQ ID No.2) ( figure 1 A). According to the enzyme cleavage site of the knockout vector pZHY988 used in the present invention, the enzyme cleavage site of BsaI was added at the primer end, and two single-stranded nucleotide sequences Os06circ02797-sgRNA-P1F were designed and synthesized (primer sequence such as SEQ ID No.3 ) and Os06circ02797-sgRNA-P1R (primer sequence such as SEQ ID No.4). It was synthesized by ...
Embodiment 2
[0123] Example 2 Verifying the Editing Efficiency of Directed Editing Expression Vectors in Protoplasts
[0124] Protoplast isolation and transformation method references for verification of editing efficiency (Tang X, Ren Q, Yang L, BaoY, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X , Zhang T, Qi Y, Zhang Y. 2019. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol J 17, 1431-1445.). Rice (Nipponbare) was cultured at 28°C in a dark environment. Cut the rice seedlings into 0.5-1.0 mm long strips with a blade. It was then quickly transferred to a 90 mm Petri dish containing 8-10 ml of enzyme solution (1.5% Cellulase R10, 0.75% Macerozyme R10, 0.6 Mannitol, 10 mM MES at pH 5.7, 10 mM Calcium Chloride and 0.1% BSA) middle. Then infiltrate under vacuum for 30 minutes. Then shake in the dark at 25° C. on a shaker at 60-80 rpm for 5-6 hours. Cell digests were subsequently filtered thro...
Embodiment 3
[0126] Rice genetic transformation mediated by embodiment 3 Agrobacterium
[0127] Agrobacterium-mediated transformation of rice references (Tang X, Ren Q, Yang L, Bao Y, Zhong Z, He Y, Liu S, Qi C, Liu B, Wang Y, Sretenovic S, Zhang Y, Zheng X, Zhang T, The experimental method disclosed in Qi Y, ZhangY. 2019. Single transcript unit CRISPR 2.0systems for robust Cas9 and Cas12 amediated plant genome editing. Plant Biotechnol J 17, 1431-1445.).
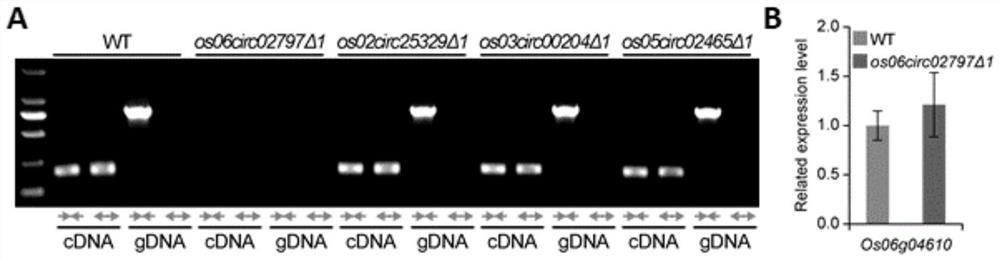
[0128] The genetic transformation steps of rice specifically include: shelling and disinfecting mature rice (Nipponbare) seeds; inoculating the sterilized seeds on N-6-D solid medium containing 0.4% gellan gum, and culturing under continuous light at 32° C. 5 days; the cultivated seeds were transformed into rice with plasmids pZJP054, pZJP053, pZJP054 and pZJP057 by Agrobacterium-mediated transformation method, and the transformed rice seeds were continuously cultured in the induction selection medium at 32°C under light for 2 weeks; th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com