Primer pair, probe and kit for detecting novel coronavirus SARS-CoV-2 and application of kit
A coronavirus, sars-cov-2 technology, applied in the direction of DNA / RNA fragments, microorganisms, recombinant DNA technology, etc., can solve the problems of backlog of suspected case samples, inability to get results quickly, inability to apply detection, etc., to achieve strong specificity , Overcoming the effects of long detection time and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
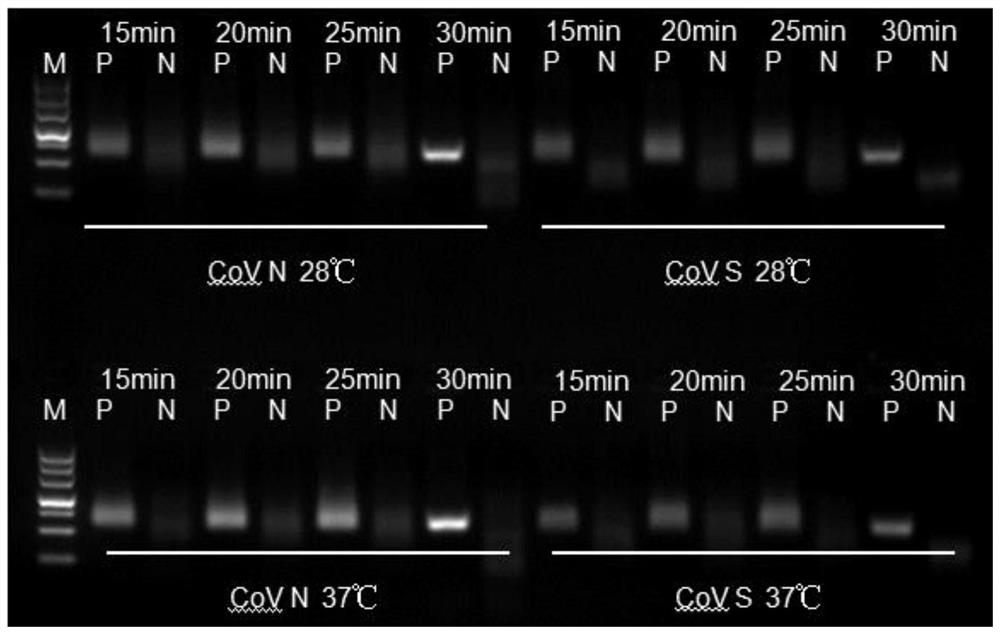
[0025] Example 1 Design of primers and probes for detection of novel coronavirus SARS-CoV-2 and RPA detection method
[0026] 1. Design of primers and probes
[0027] Through comprehensive analysis and comparison of the published full sequence of the new coronavirus SARS-CoV-2 including 10 genes, the N and S gene sequences were selected for primer probe design. Through screening, two sets of primer probe sets listed in Table 1 below are preferably selected, wherein the first set is the upstream primer shown in SEQ ID NO.1 and the downstream primer shown in SEQ ID NO.3 designed for the N gene sequence , and the probe shown in SEQ ID NO.5; the second group is the upstream primer shown in SEQ ID NO.2, the downstream primer shown in SEQ ID NO.4, and the downstream primer shown in SEQ ID NO.6 designed for the S gene sequence probe.
[0028] Table 1 Sequences of primers and probes for the detection of novel coronavirus SARS-CoV-2
[0029]
[0030] Preferably, the 29th base of ...
Embodiment 2
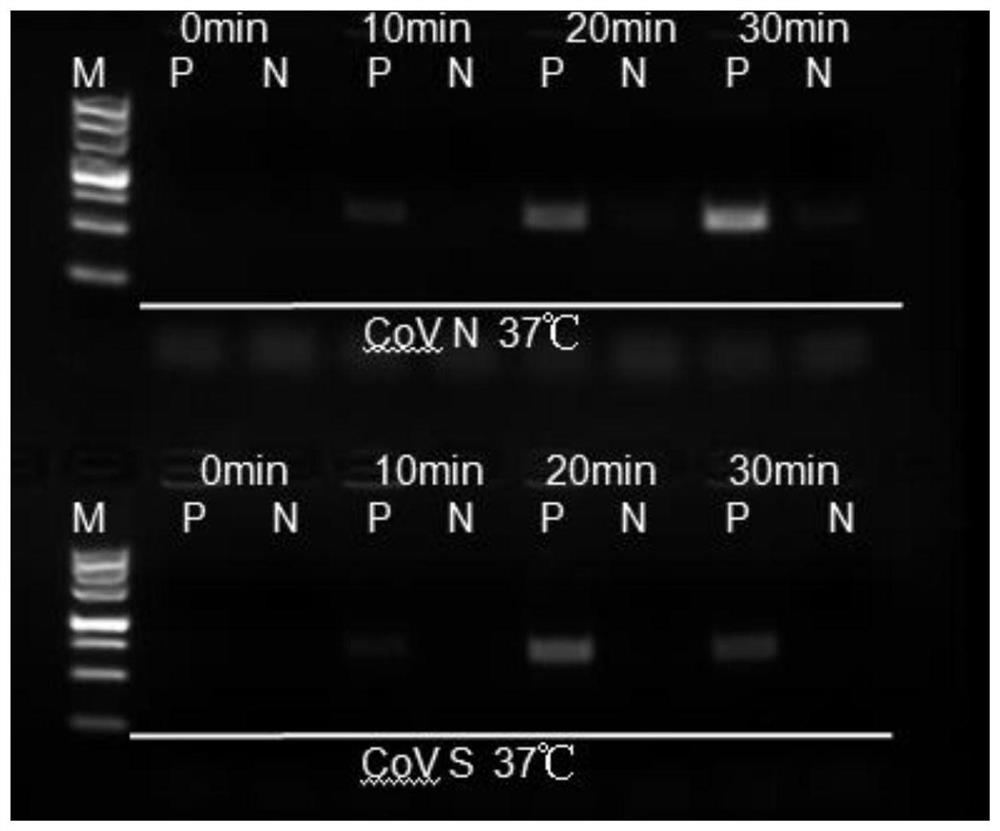
[0038] Example 2 Detection method of rt-RPA for novel coronavirus SARS-CoV-2
[0039] 1) Take out the required components of the TAEXO02KIT kit 30 minutes in advance, thaw at room temperature, and mix by shaking.
[0040] 2) The primers and probes were dissolved in water, and the final concentration was 10 μM.
[0041] 3) Add 29.4 μL A buffer, 3 μL upstream primer, 3 μL downstream primer, 2 μL probe, 5.1 μL RNase free H to each dry powder reaction tube 2 O, invert the reaction tube 8-10 times and mix thoroughly.
[0042] 4) Add 5 μL RNA template and 2.5 μL B buffer to the reaction tube, invert up and down 8-10 times to mix, then centrifuge the reaction solution to the bottom of the tube briefly, and then immediately put the reaction tube into the fluorescence detection equipment (such as constant temperature expansion) increaser, real-time quantitative PCR, etc.).
[0043] 5) The constant temperature is 37°C; the fluorescence signal is collected every 30s (the selection of ...
Embodiment 3
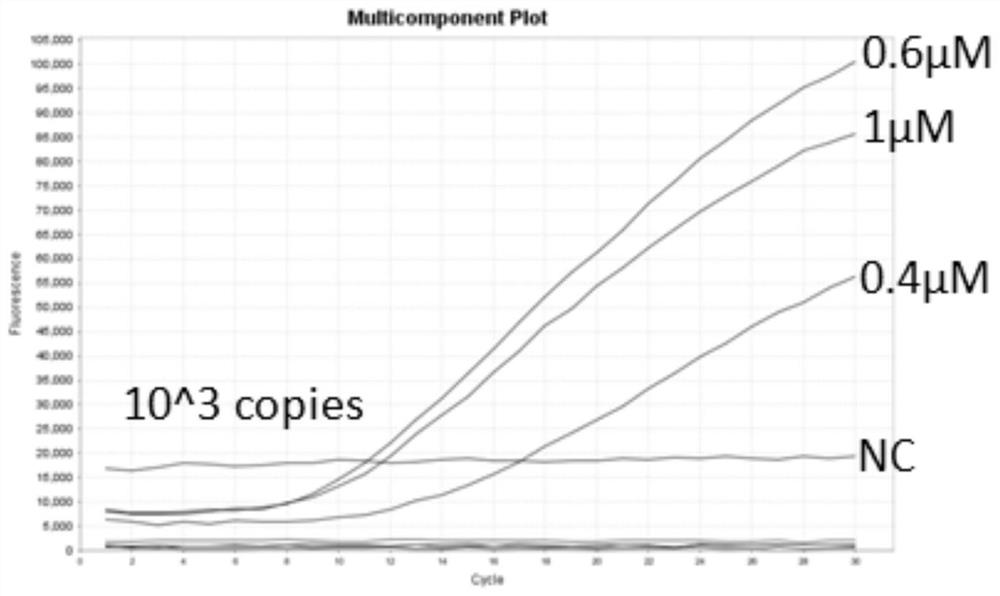
[0045] Example 3 Optimization of Primer Concentration
[0046] with 10 3 Copie / μL plasmid standard is used to optimize the primer concentration, and the specific implementation is as follows:
[0047] 1) Take out the required components of the TAEXO02KIT kit 30 minutes in advance, thaw at room temperature, and mix by shaking.
[0048] 2) The primers and probes were dissolved in water, and the final concentration was 10 μM.
[0049] 3) Set up four experimental tubes, add 29.4 μL A buffer and 2 μL probe to each dry powder reaction tube, add 2 μL upstream primer and 2 μL downstream primer to 0.4 μM group, and add 3 μL upstream primer and 3 μL downstream primer to 0.6 μM group, 1.0 5μL upstream primer and 5μL downstream primer were added to μM group, NC group was a negative control, and an appropriate amount of RNase free H was used. 2 O Make up the system to 42.5 μL, invert the reaction tube up and down 8-10 times, and mix well.
[0050] 4) Add 5 μL RNA template and 2.5 μL B bu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com