Method to enhance the transcription regulation of SUPT4H on genes containing repetitive nucleotide sequences
A repeat sequence, nucleotide technology, applied in gene therapy, genetic engineering, biochemical equipment and methods, etc., can solve problems such as toxicity, defective genes, and unclear molecular mechanisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Embodiment 1. Experimental material and method:
[0068] A. Yeast strains and plastids
[0069] Yeast strains with gene deletion were generated by the one-step gene replacement method and confirmed by PCR (Liu et al., 2012). The ORFs of SPT4 and RNH1 in W303-1A cells were replaced by KanMX and NatMX, respectively To obtain SPT4Δ (SPT4 gene knockout (gene knock-out, gene knockout)) or RNH1Δ (RNH1 gene knockout) strains.
[0070] Plastids with different lengths and nucleotide sequences, including repeat sequences CA, CAA, CAG, CAAC and CAACCA, were prepared by the technical method disclosed in (Liu et al., 2012). Using yeast chromosomal DNA as a template, the full-length TOP1 gene coding sequence and the N-terminal part of SEN1 (encoding amino acid residues 1 to 1017) were amplified by PCR, and the amplified product of TOP1 (by SmaI-TOP1F and TOP1-NotI R primer) and the amplified products of SEN1 (via SmaI-SEN1F and SEN1-NotI R primers) were inserted into the gene expre...
Embodiment 2
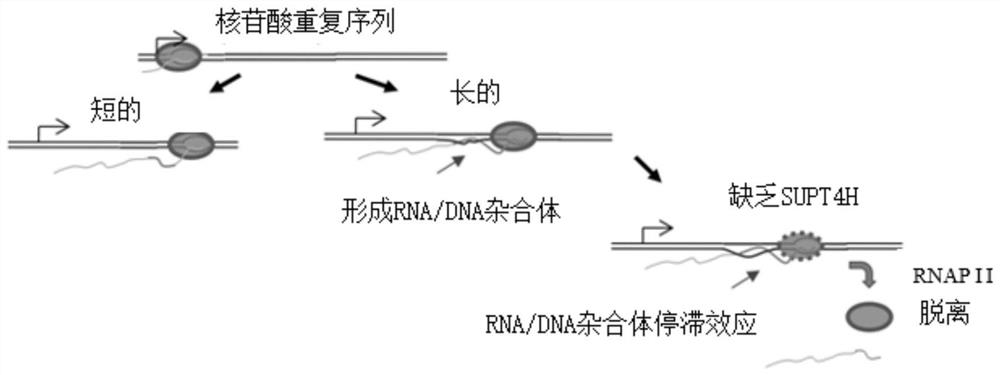
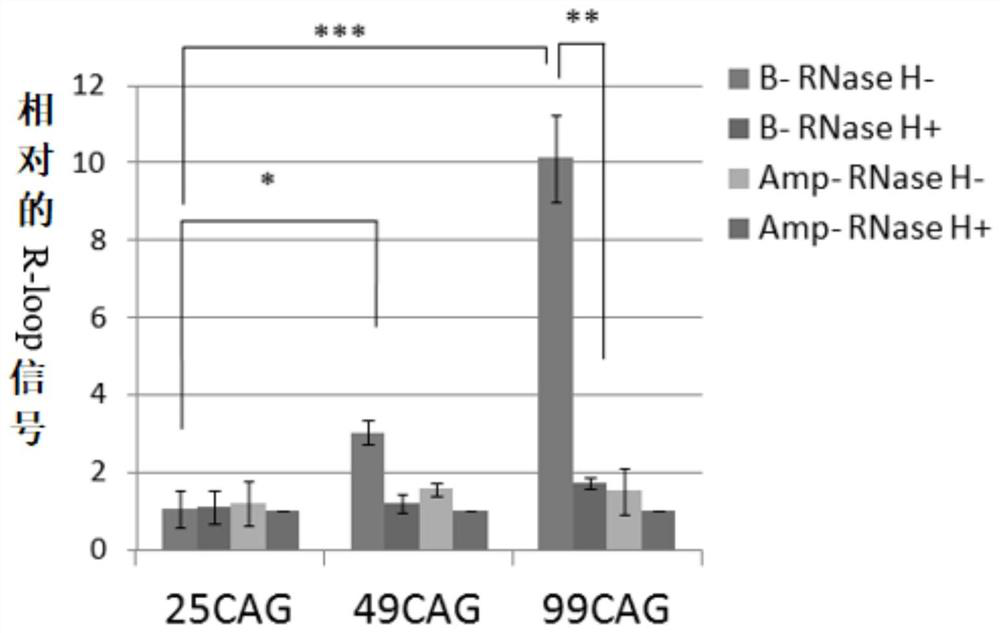
[0097] Example 2: The formation of R-loops in transcription is affected by the number of CAG repeats, but not by SPT4 in yeast cells.
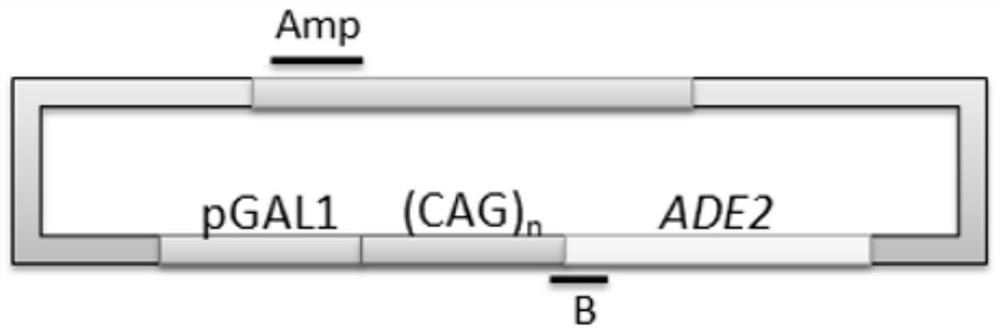
[0098] Figure 2A Amplicons on (CAG)n-ADE2 plasmids by primer set B and primer set Amp are shown. (CAG)n-ADE2 comprises the ADE2 reporter gene fused to repeats of 10, 25, 49 or 99 CAGs. The fusion gene was under the control of the GAL1 promoter (pGAL1) to induce (CAG)n-ADE2 expression by culturing the cells in a medium containing 2% galactose. Primer set B amplifies the last 3 CAG repeat sequences of DNA to the position of repeat unit 200bps. Primer set Amp amplification amp R The gene is used as the control group, and the primer group Amp does not produce R-loops. ; Figure 2B R-loops signal on (CAG) n-ADE2 during transcription analysis by RNA / DNA Hybridization Precipitation (DIP) method is shown. use (CAG) 10 -ADE2, (CAG) 25 -ADE2 or (CAG) 49 -ADE2 was introduced into wild-type (WT) cells grown in galactose-containing medium and the...
Embodiment 3
[0100] Example 3: In Spt4-deficient cells, increasing the formation of R-loops reduces the transcriptional effect of CAG repeats.
[0101] Figure 3A Shows signaling of R-loops in wild-type (WT), RNH1Δ(Δ1), SPT4Δ(ΔS), and RNH1ΔSPT4Δ double-deleted (Δ1ΔS) cells, feeding these cells into (CAG) 99 -ADE2, grown in a medium containing galactose, and then analyzed for R-loops signal by DIP. The experimental data is compared with the Amp-RNase H+ sample after normalization, such as Figure 2B Said (N=3; * represents P Figure 3B Shows the expression level of (CAG)n-ADE2 mRNA measured by Northern blotting in WT, Δ1, ΔS and Δ1ΔS cells. Will (CAG) 25 -ADE2 or (CAG) 99 - ADE2 was transfected (transformed) into cells, and then RNA was obtained for quantitative analysis. After correction, as described in Figure 2D, (CAG) in WT cells 99 - The expression level of ADE2 mRNA was set as 100% (N=3). Figure 3C It is a schematic diagram of the amplicon formed by primer set A and primer set ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com