In-vitro screening method of nucleic acid aptamer, nucleic acid aptamer and kit for detecting target molecule
A nucleic acid aptamer, in vitro screening technology, applied in the biological field, can solve the problems of the existence of PCR product fidelity, nucleic acid aptamers need to be studied, monomer synthesis difficulties, etc., to achieve easy screening, simple synthesis method, and applicability wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] 1. Construction of PS-random library
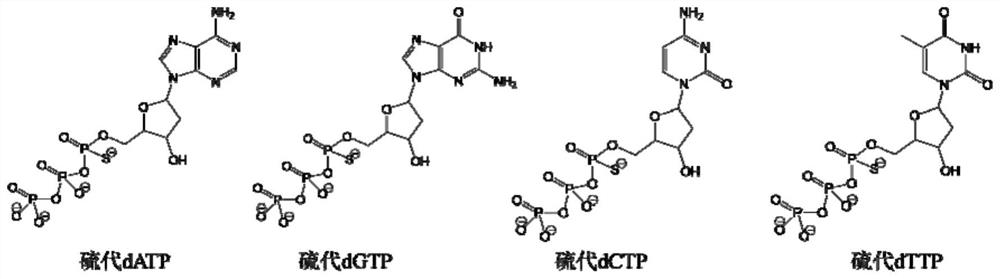
[0043] First obtain a random library through solid-phase synthesis, the sequence is ATACCAGCTTTCTGTTCT-N30-AGATAGTAAGTGCAATCT, and synthesize B-DNA, its sequence is: biotin-AAAAAAAAAAAAAGAACAGAAAGACC, the random library can be hybridized with the B-DNA through the middle GGTCTTTCTGTTCT sequence, Then connect to the avidin-modified magnetic ball through the biotin on the B-DNA. At the same time, synthesize primers for PCR, the forward primer is AP1: 5'-ATACCAGCTTATTCAATT-3', the reverse primer is TER-AP2: 5'-A20-Spacer18-AGATTGCACTTACTATCT-3', and the thio monomer is selected from thio dATP, other monomers are natural monomers dTTP, dGTP, dCTP. Add 2.5 μl AP1, 2.5 μl TER-AP2, 1 μl 10 mM sulfo-dATP, 0.5 μl dTTP, 0.5 μl dGTP, 0.5 μl dCTP, 41.5 μl HO to a 200 μl PCR tube 2 O. 0.5 μl Phusion enzyme and 0.5 μl random library template. Place the above PCR system on a PCR machine, denature at 98°C for 3min, denature at 98°C for 20s, ann...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com