Micro nucleic acid combined amplification testing method and kit
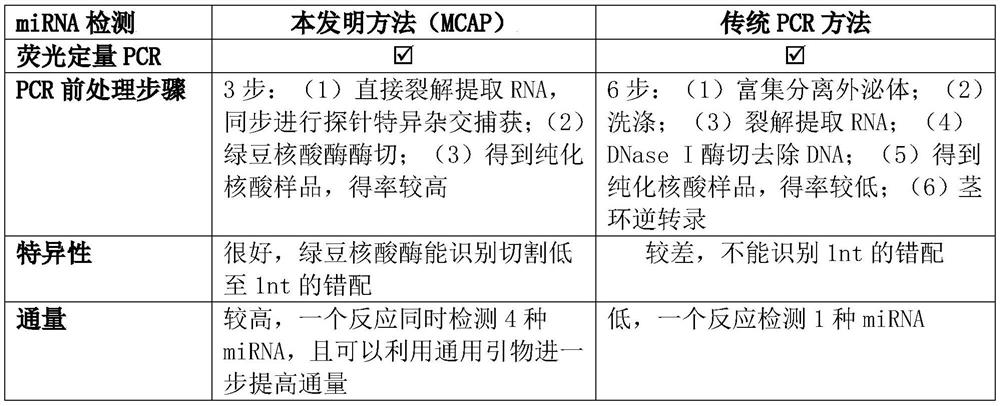
A technology of tiny nucleic acid and detection method, applied in the field of biochemistry, can solve the problems of low throughput, poor repeatability, complicated operation, etc., and achieve the effect of reducing complexity, enhancing specificity and stability, and enhancing sensitivity and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
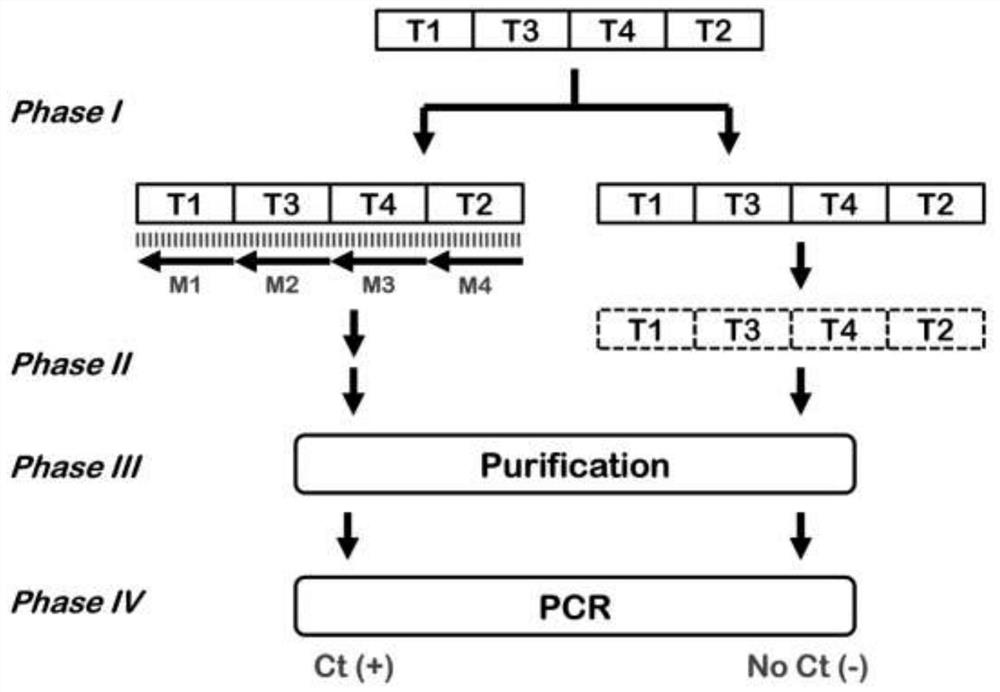
[0024] Example 1: Using the method and kit to detect oligonucleotide probes mixed in Escherichia coli samples The composition of the kit for this method is as described in the text of the manual. We synthesized a template probe TSNOT that fully binds to four human miRNAs: miR-191, miR-93, miR-16, and miR-3662. The sequence is: CAGCTGCTTTTGGGATTCCGTTGCGCCAATATTTACGTGCTGCTACATCAGTCACTACTCATCATTTTCGTTTCACGACAAGCACGTCCATC, where the complementary sequences of miR-191 and miR-93 The complementary sequences of miR-16 and miR-3662 are located at the 5' and 3' ends of TSNOT respectively, and the complementary sequences of miR-16 and miR-3662 are connected in the middle of TSNOT, and the four sequences are connected continuously without any additional gap. As a format validation for co-detection of oligonucleotide probes spiked into E. coli samples. The oligonucleotide probe sequences were identical to the above four miRNAs: OLI-191, OLI-93, OLI-16, OLI-3662, and the sequences were: CA...
Embodiment 2
[0025] Example 2: Using this method and kit to detect 4 miRNAs in human lung cancer A549 cells
[0026] The composition of the method kit is as described in the text of the specification. The sequences of template probe TSNOT, primers PMF191 and PMR93 are as described in Example 1. We further use template probe TSNOT, primer PMF191, primer PMR93 system, with 20μl 10 5 / μl E. coli samples for negative samples, to 20μl 10 4 The human lung cancer A549 cell sample / μl is a positive sample, and the joint detection of four human miRNAs, miR-191, miR-93, miR-16, and miR-3662, is performed. The operation steps were the same as those described in Example 1, and the detection results: negative samples had no amplification, and positive samples had a Ct value of 26.77, indicating that the system can be used for joint detection of the four target miRNAs.
Embodiment 3
[0027] Example 3: Using this method and kit to detect 4 miRNAs in the culture medium of human lung cancer A549 cells
[0028] Cancer cells can actively secrete miRNAs to the surrounding environment through exosomes and microvesicles. These extracellular miRNAs can stably exist in body fluids or culture fluids for a long time due to the protection of the coating membrane. We cultured A549 cells continuously for 3 days, so that the growth density was close to 100%, and then collected 10 μl of culture medium, and used this method and kit to detect four human miRNAs, miR-191, miR-93, miR-16, and miR-3662. joint detection. The composition of the method kit is as described in the text of the specification. Template probe TSNOT, primers PMF191, PMR93 sequences, and operation steps are as described in Example 1. We further use template probe TSNOT, primer PMF191, primer PMR93 system, with 20μl 10 5 / μl of E. coli samples for negative samples, to 20μl10 4 The human lung cancer A54...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com