Modified nucleic acid inhibiting micro rna, and use thereof
A nucleic acid and nucleotide technology, which is applied in the field of modified nucleic acid to inhibit microRNA, can solve the problems of adverse effects, inability to distinguish normative targets from non-normative targets, etc., and achieve the effect of preventing competitive inhibition and effectively inhibiting miRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
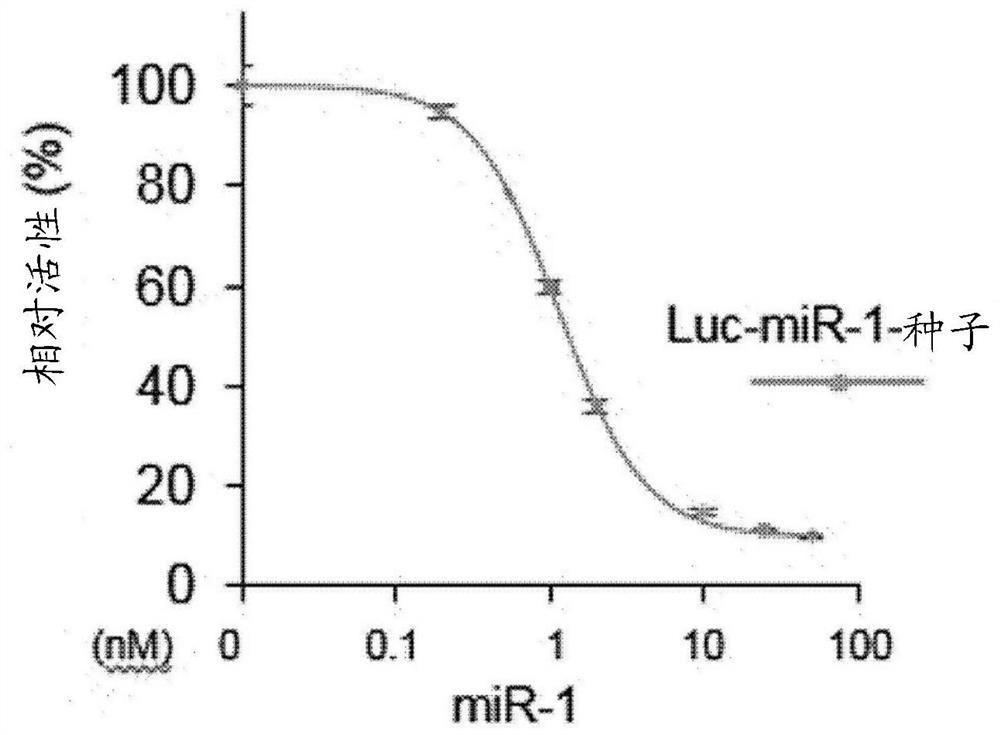
[0059] There is no limitation on the type of miRNA according to the present invention, and according to an exemplary embodiment of the present invention, the miRNA may be miRNA-1.
[0060] In the present invention, the miRNA target site sequence may include at least 6 nucleotides, preferably 21 or less nucleotides, most preferably 6 to 8 nucleotides, but the present invention is not limited thereto.
[0061] Since the miRNA actually only recognizes at least 6 base pairs in positions 1 to 8 from its 5' end, even a sequence complementary only to the base sequence in positions 1 to 8 can sufficiently bind to the miRNA, and When the miRNA target site sequence consists of 6 to 8 nucleotides, only the canonical target recognition of the miRNA can be competitively inhibited.
[0062] In the present invention, the miRNA-inhibiting modified nucleic acid may contain at least two miRNA target site sequences consecutively, and is better because it contains two or more miRNA target site se...
Embodiment 1
[0095] Example 1. Demonstration of the effect of inhibiting the canonical seed-binding function of miRNAs by tandem target miRNA inhibitors
[0096] 1-1. Method for preparing tandem target miRNA inhibitors
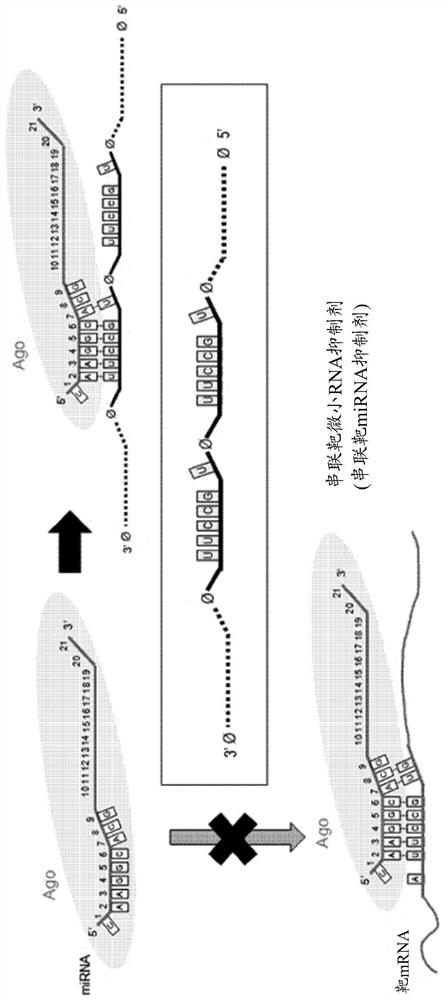
[0097] Such as figure 1 As shown, 5'-UAAGGCACG-3' is a base sequence from 1 to 8 positions from the 5' end of the base sequence of the miRNA that binds to the Argonaute protein, and is the seed base of the corresponding miRNA (for example, miR-124) base sequence, and the miRNA uses this sequence to bind complementary to the target mRNA. Here, when a tandem target miRNA inhibitor (a modified nucleic acid that inhibits miRNA) is prepared by introducing two or more short 8-nt target sequences capable of complementary pairing with the miRNA seed, the miRNA seed (5'-UAAGGCACG- 3') and the 5'-UGCCUU-3' sequence of the tandem target miRNA inhibitor are fully complementary, thereby relatively inhibiting the base pairing with the actual target mRNA and inhibiting the canonical ...
Embodiment 2
[0112] Example 2. Discovery and validation of non-canonical target sites that allow wobble base pairing in miRNA seed regions
[0113] 2-1. Ago HITS-CLIP method to detect non-canonical G:A wobble pairing sites of miRNAs expressed in the cerebral cortex
[0114] As a method for analyzing miRNA targets at the transcriptome level, an experimental method called AgoHITS-CLIP has been developed and widely used. AgoHITS CLIP analysis is a method in which RNA in cells is induced by irradiating cells or tissue samples with ultraviolet light. Covalently bonded to Argonaute proteins, the resulting RNA-Argonaute complexes were isolated by immunoprecipitation using antibodies specific for Argonaute, and the isolated RNA was analyzed by next-generation sequencing. The base sequence data thus obtained can not only identify target mRNAs of miRNAs through bioinformatics analysis, but also precisely analyze their binding sites and sequences. AGO HITS-CLIP was first applied to mouse cerebral ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| strength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com