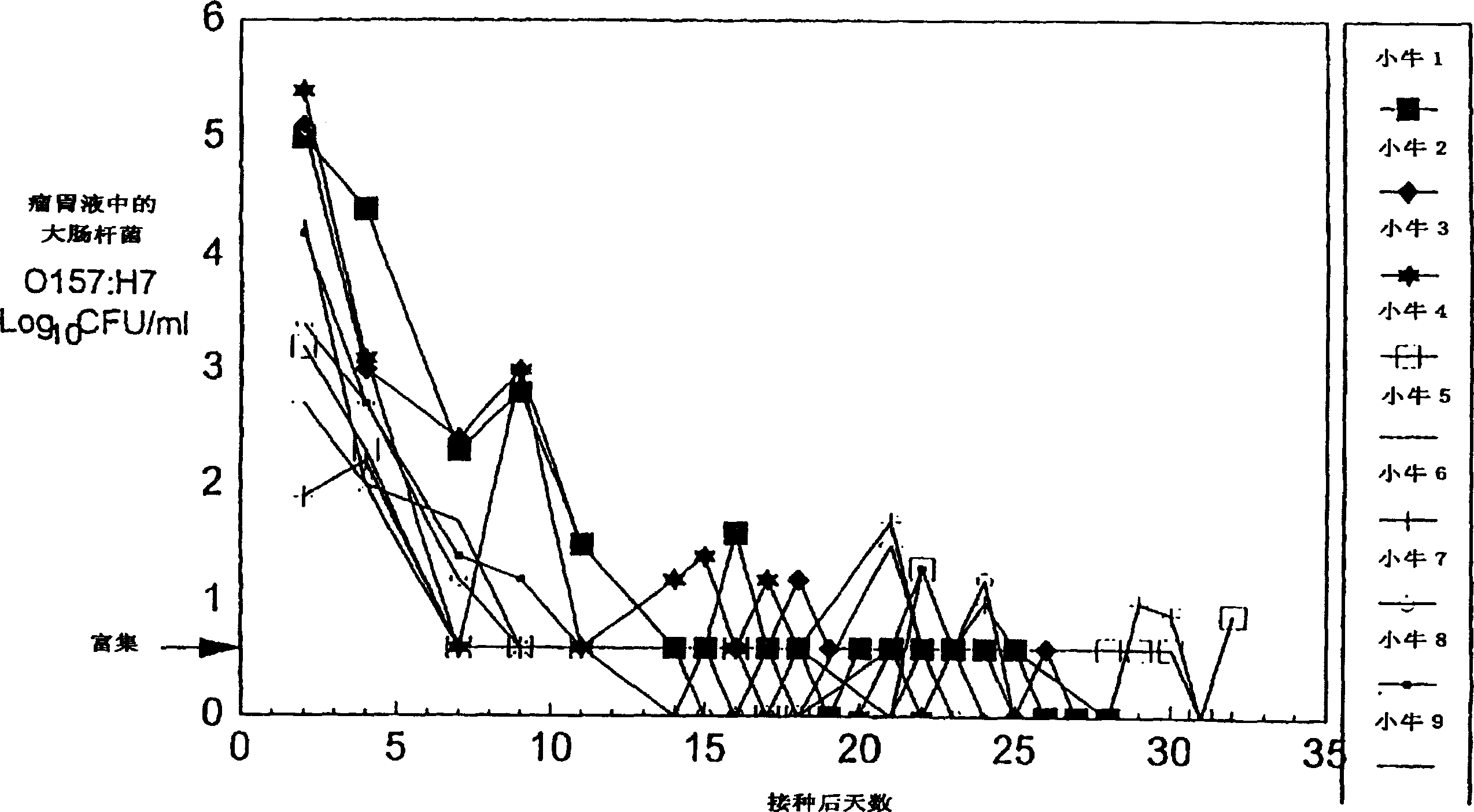
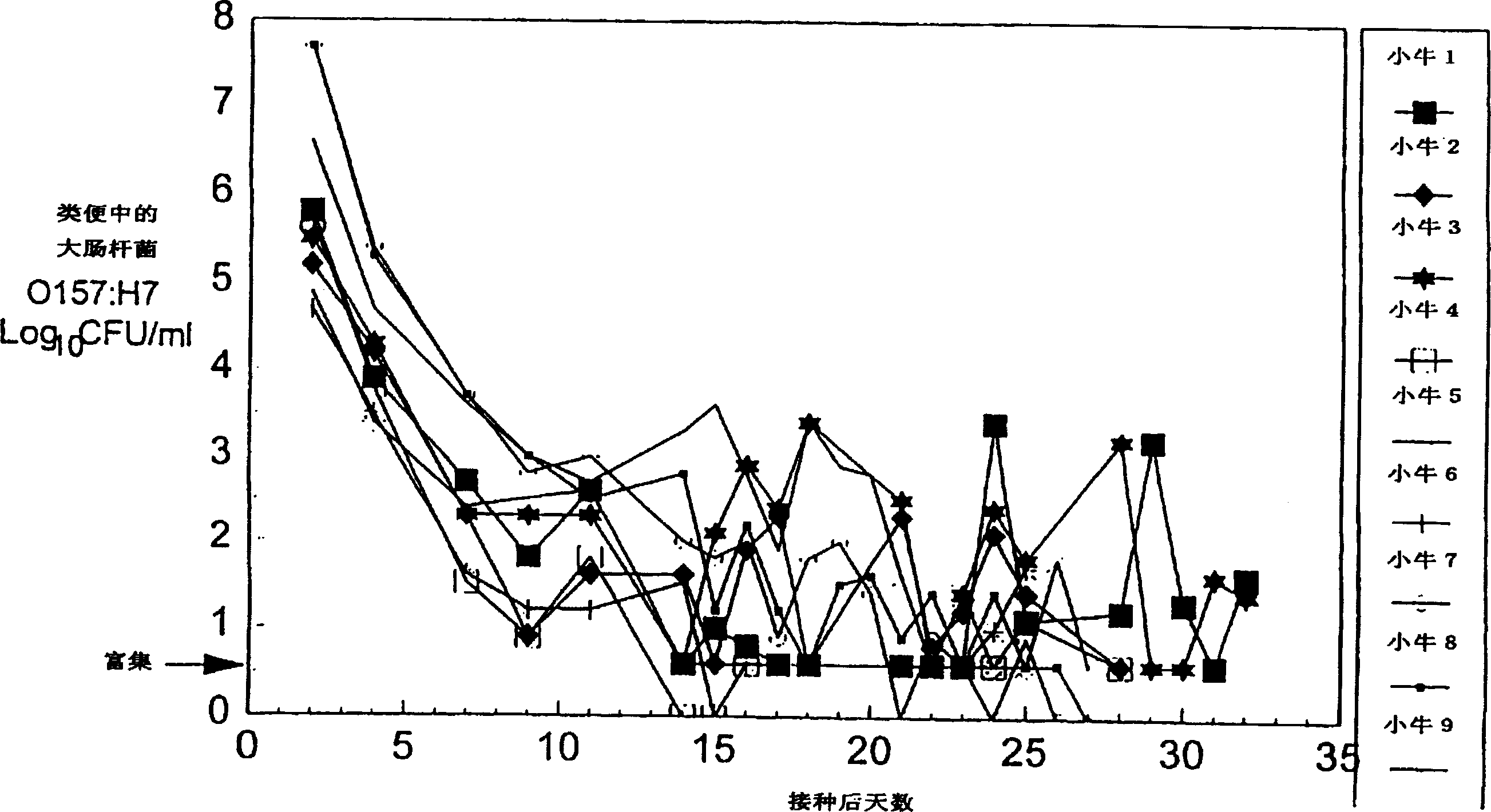
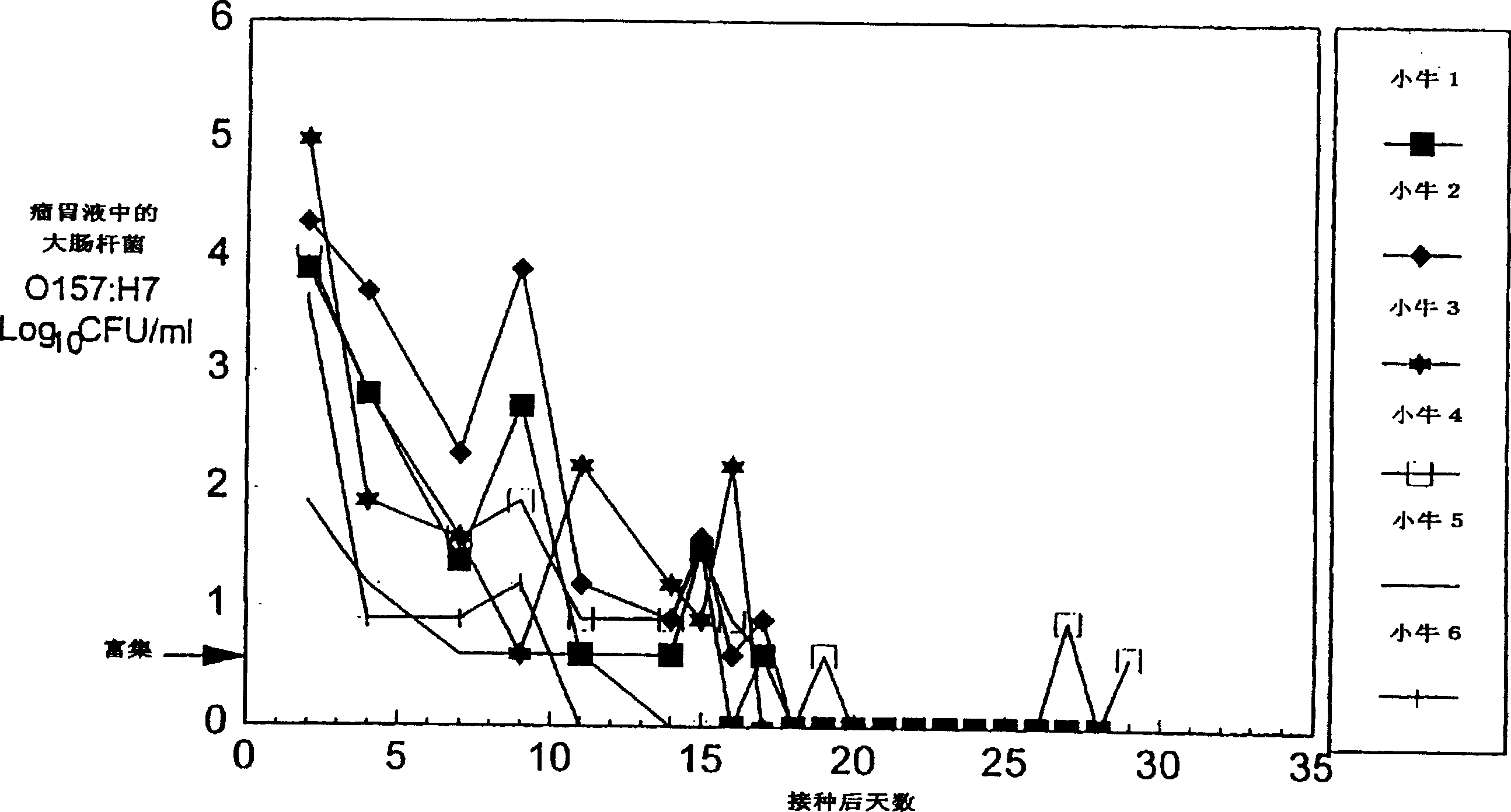
Control of enterohemorrhagic E Coli 0157:H7 of cattle with probiotic bacteria
A technology of Escherichia coli and O157, which is applied in the direction of antibacterial drugs, bacteria, and medical raw materials derived from bacteria, and can solve the problems of biological control that have not yet been obtained
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1 Isolation of Prior Antibiotic Bacteria
[0032] Isolation of pre-antibiotic-resistant bacteria from livestock feces or livestock gastrointestinal tissue (small intestine and colon). Fecal samples were collected from livestock that were confirmed negative for E. coli O157:H7 by fecal testing. Use 0.1M phosphate buffer (pH7.2) [PBS] to make a 1:10 serial dilution of the stool sample, and take 0.1ml of each dilution and plate it on a sorbitol MacConkey agar (SMA) plate. Plates were incubated at 37°C for 16 hours until 10 colonies were randomly selected and each colony was transferred to a tube containing 10 ml of Tryptic Soy Broth (TSB) (BBL, Becton Dickinson, Cockeysville, MD). The culture solution was incubated at 37°C for 16 hours. Tissue samples (1 g each) were homogenized at 9,500 rpm for 1 minute (Ultra-Turrax T25 homogenizer, Janke & Kunkel IKA-labortechnik, Germany), and then 0.1 ml each was spread on the surface of an SMA plate. Plates were incubated ...
Embodiment 2
[0033] Example 2 Screening of cultures resistant to Escherichia coli O157:H7 properties
[0034] The culture supernatants were screened against E. coli O157:H7 metabolites with a mixture of 5 strains of E. coli O157:H7, including 932 (human isolate), C7927 (human isolate), E009 (meat isolate), E0018 (livestock isolate) and E0122 (livestock isolate). Roughly equalize about 10 per plant 7 E. coli O157:H7 in 0.1 ml was surface plated on duplicate plates of SMA and TSA. A disc (12 mm in diameter) was placed on the surface of each SMA and TSA plate, and 0.1 ml of the filter-sterilized supernatant of a single culture was added to the disc surface. Additionally, disks containing 70% ethanol (positive control) and PBS (negative control) were added to each plate. Cultures were incubated at 37°C for 18 hours and zones of inhibition were observed. A clear band above 1mm was regarded as a positive reaction.
Embodiment 3
[0035] Example 3 Preparation of Escherichia coli O157:H7 culture
[0036] Use the same 5-strain mixture as above. To facilitate enumeration of these bacterial isolates, tolerance to nalidixic acid (50 μg / ml) was induced in these strains. Transfer each nalidixic acid-resistant Escherichia coli O157:H7 strain to 10 ml of tryptic digested soybean broth (TSB) containing nalidixic acid (50 μg / ml), and incubate at 37°C with shaking (150rpm) for 16- 18 hours. Transfer 2 ml of the suspension of each isolate into 300 ml of TSB. After incubation at 37°C for 16-18 hours, the bacteria were pelleted by centrifugation (4,000×g, 20 minutes), and washed 3 times with PBS. Add PBS in the bacterium of precipitation, its amount is to obtain optical density (O.D.) 0.5 (10 8 CFU / ml) required amount. Before the oral inoculation of calves, 5 strains of Escherichia coli O157:H7 isolates (2 × 10 9 CFU) was mixed in 250ml of 2% sterilized skimmed milk. The number of bacterial colonies was determi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com