Method for detecting uracil-DNA glycosylase activity by using fluorescent probe
A technology of glycosylase activity and fluorescent probe, which is applied in the field of fluorescent probe detection of uracil-DNA glycosylase activity, can solve problems such as incorrectness and high risk of cross-contamination, and achieve high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
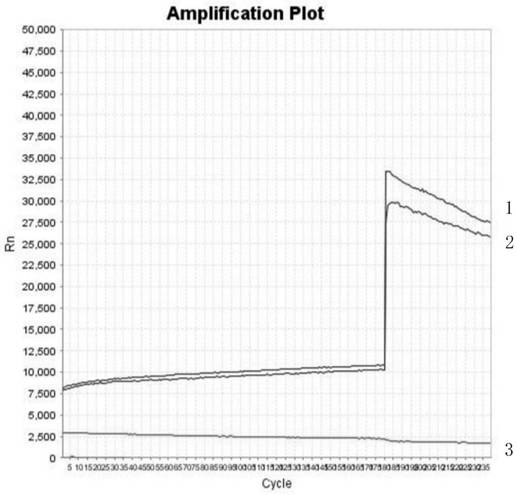
[0022] like figure 1 A method for detecting uracil-DNA glycosylase activity with a fluorescent probe is shown, the method is to mix a polymerase chain reaction buffer solution and a fluorescent resonance energy transfer based method in a real-time polymerase chain reaction instrument Fluorescent DNA probe, uracil-DNA-glycosylase; take 10 µl of polymerase chain reaction buffer.
[0023] The reaction solution is incubated at 25°C-37°C for 30 minutes, at this time, the fluorescent DNA probe nucleic acid-labeled fluorescent dye based on fluorescence resonance energy transfer is excised. Collect pyrimidine-DNA glycosylase activity data (that is, Rn fluorescence value data) in the fluorescence measurement channel at a wavelength of 520 nm every 10 seconds, and then heat at 95°C for 2 minutes to inactivate uracil-DNA-glycosylase. Incubate at 25°C-37°C for 10 minutes to degrade fluorescent DNA probes based on fluorescence resonance energy transfer. Collect pyrimidine-DNA glycosylas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com