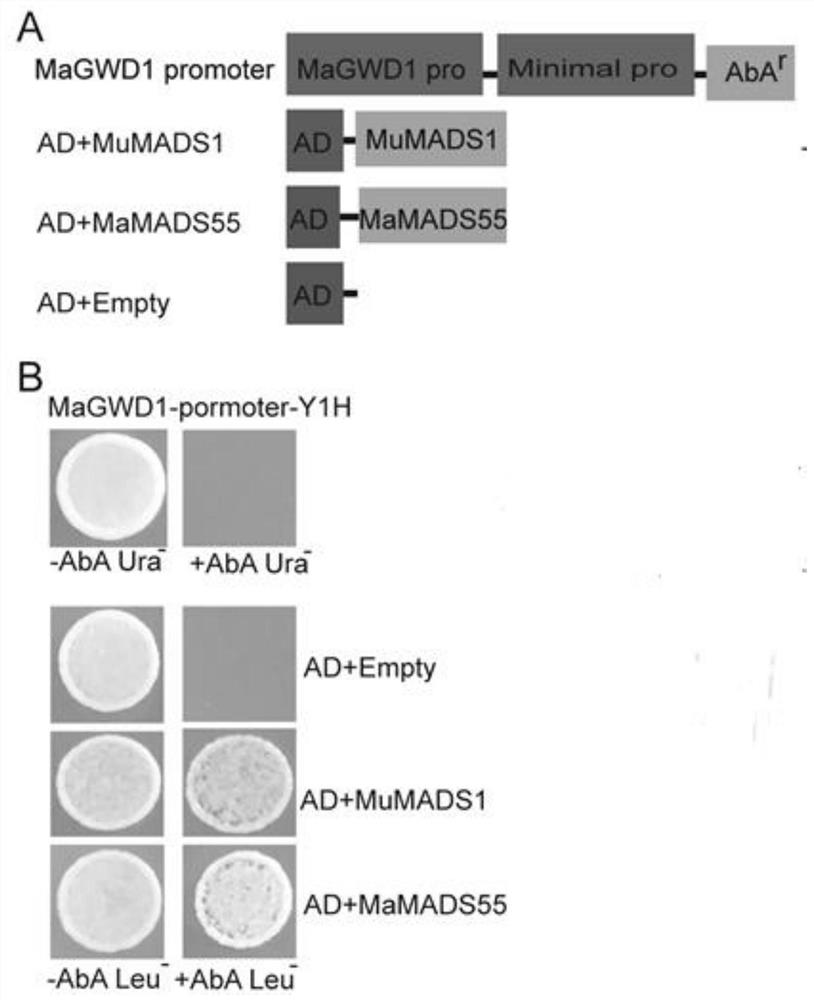
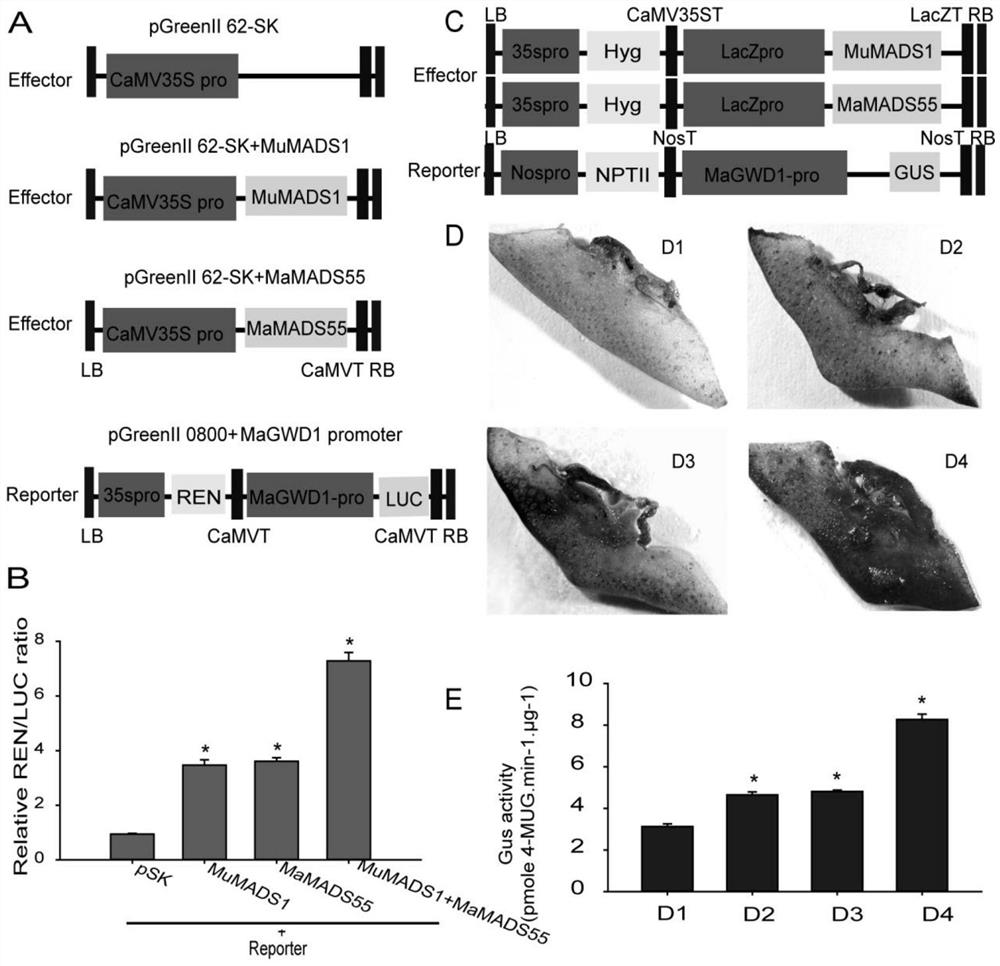
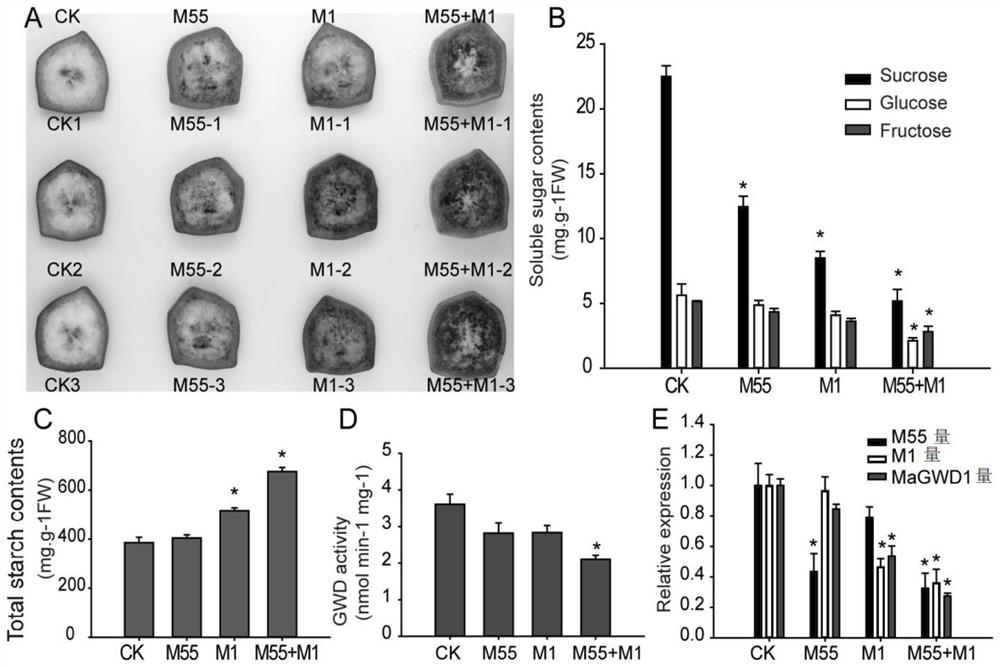
Application of interaction of banana MuMADS1 and MaMADS55 in regulation and control of MaGWD1 gene expression
A gene expression and gene technology, applied in the biological field, to improve quality and promote up-regulation of expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] Referring to the accompanying drawings, the present invention will be further described in conjunction with specific embodiments, so as to better understand the present invention. If no specific technique or condition is indicated in the examples, it shall be carried out according to the technique or condition described in the literature in this field or according to the product specification. The reagents or instruments used were not indicated by the manufacturer, and they were all commercially available conventional products.
[0030] 1 Experimental materials
[0031] (1) Bananas (M. acuminata L. AAA group cv Brazilian) were obtained from the banana plantation of the Institute of Tropical Biotechnology (20N, 110E, Chiang Mai, Hainan Province, China), and were transported to the laboratory immediately after harvest. Select a healthy fruit comb and divide it into individual fruit fingers. Select intact and undamaged fruit fingers and sterilize the surface with 0.1% sod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com