Primer and probe combination product for detecting SARS-CoV-2
A combined product and primer-probe technology, which is applied in the field of molecular biology, can solve problems such as the lack of widespread promotion and application, the sensitivity and specificity gap of constant temperature amplification technology, and achieve high-efficiency constant temperature nucleic acid amplification with low hardware requirements. , Conducive to the widespread promotion of technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
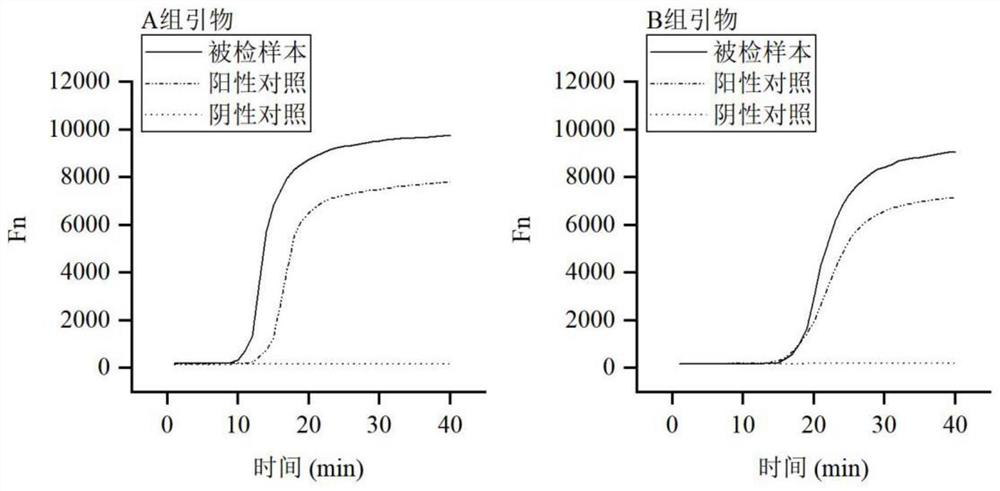
[0068] Embodiment 1 Establishment of a SARS-CoV-2 virus fluorescence detection kit detection method
[0069] Combined with the recommendations in the guidelines and the analysis of the conservation of the SARS-CoV-2 genome, the N gene was selected for primer and probe design. The designed primers and probes are as follows:
[0070] Group A:
[0071] Primer F3: 5'-AACACAAGCTTTCGGCAG-3' (SEQ ID NO: 1)
[0072] Primer B3: 5'-GCGTCAATATGCTTATTCAGC-3' (SEQ ID NO: 2)
[0073] Primer FIP: 5'-TGCGGCCAATGTTTGTAATCAGCCAAGGAAATTTTGGGGAC-3' (SEQ ID NO: 3)
[0074] Primer BIP: 5'-TCAGCGTTCTTCGGAATGTCGCTGTGTAGGTCAACCACG-3' (SEQ ID NO: 4)
[0075] Primer LF: 5'-TTCCTTGTCTGATTAGTTC-3' (SEQ ID NO: 5)
[0076] Primer LB: 5'-TGGCATGGAAGTCACACC-3' (SEQ ID NO: 6)
[0077] Primer probe: 5'-FAM-AATTGCACAATTT[rG]CCCCCAGCGCT-BHQ1-3' (SEQ ID NO: 7)
[0078] Group B:
[0079] Primer F3: 5'-TGGCTACTACCGAAGAGCT-3'
[0080] Primer B3: 5'-TGCAGCATTGTTAGCAGGAT-3'
[0081] Primer FIP: 5'-TCTGGCCCAGT...
Embodiment 2
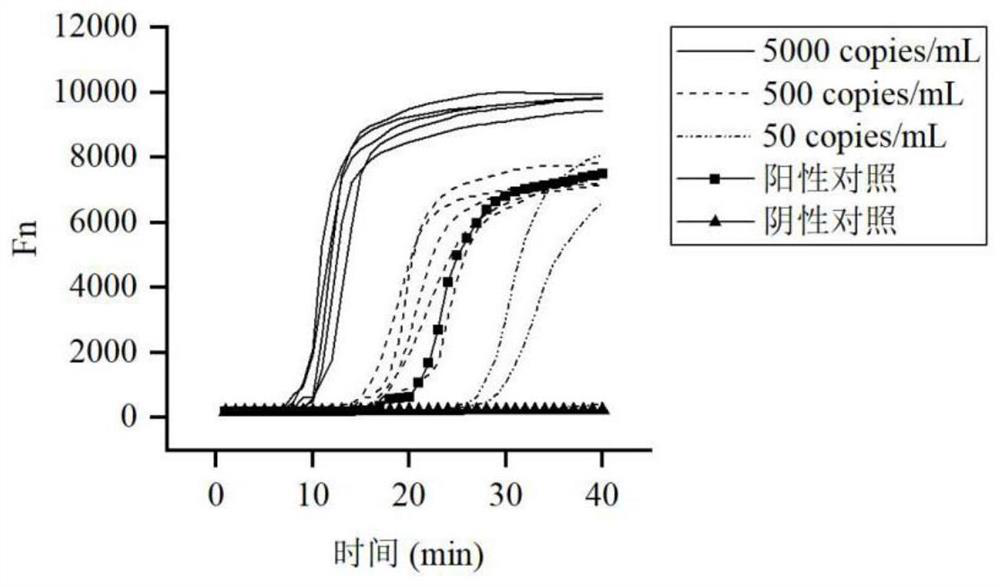
[0106] Example 2 Sensitivity test of SARS-CoV-2 virus fluorescence method detection kit
[0107] The SARS-CoV-2 genome standard substance was diluted to 5000copies / mL, 500copies / mL, and 50copies / mL respectively according to the N gene, and the sensitivity of the kit was tested.
[0108] The specific operation is as follows:
[0109] Standard substances of different concentrations, as well as negative and positive controls were taken for detection, and each concentration gradient of the standard substance was tested 5 times respectively, and the results were recorded.
[0110] The result is as figure 2 shown. The positive and negative control test results were normal. All concentrations of 5000copies / mL and 500copies / mL are detected, and 50copies / mL is detected with probability. According to the conversion of 10μL sample volume, concentrations above 5copies / reaction can be detected stably, and 0.5copies / reaction is affected by sampling with a probability of detection.
Embodiment 3
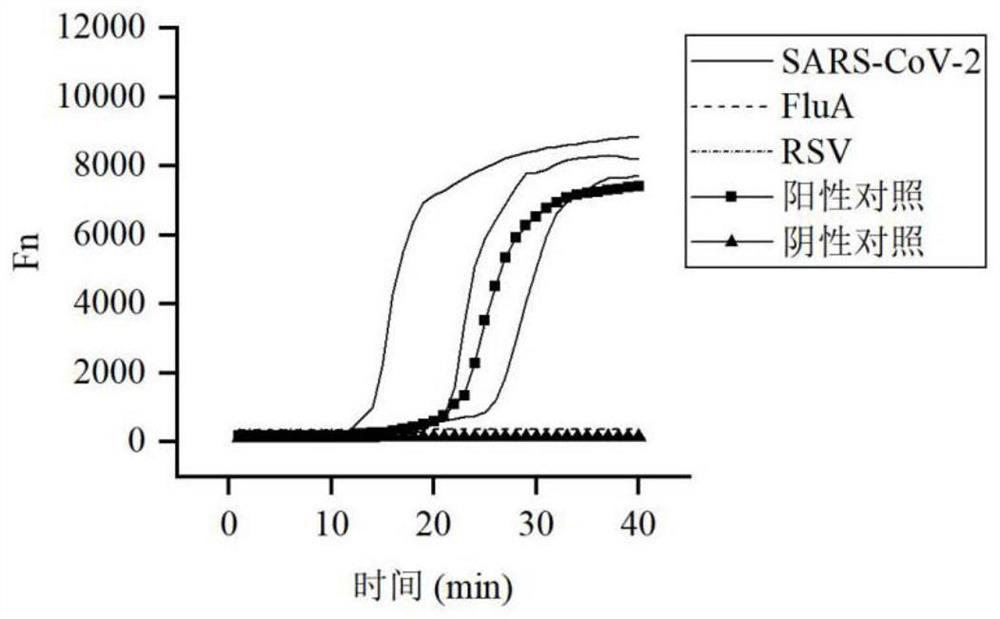
[0111] Example 3 SARS-CoV-2 virus fluorescence method detection kit specificity test
[0112] Three cases of SARS-CoV-2, one case of influenza A virus (Influenza A virus, FluA), and one case of respiratory syncytial virus (Respiratory Sycytial Virus, RSV) were collected clinically, a total of 5 cases were verified by fluorescent quantitative PCR as The corresponding virus-positive samples were tested to test the specificity of the kit.
[0113] The specific operation is as follows:
[0114] Take 5 samples and negative and positive controls respectively for detection, and record the results.
[0115] The result is as image 3 shown. Three cases of SARS-CoV-2 were positive, one case of FluA and one case of RSV were negative, and the negative and positive controls were normal. Show the specificity of the detection result of the kit of the present invention.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com