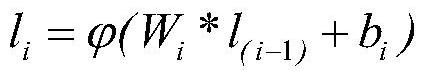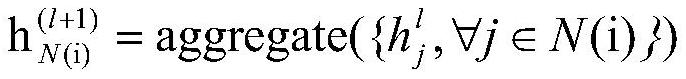Molecular structure prediction method based on graph convolutional network
A molecular structure and prediction method technology, applied in the field of machine learning, can solve the problems of high cost, tediousness, and the inability to balance the relationship between calculation accuracy and time cost, and achieve the effect of optimal time and good prediction structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] Below in conjunction with accompanying drawing, describe technical scheme of the present invention in detail:
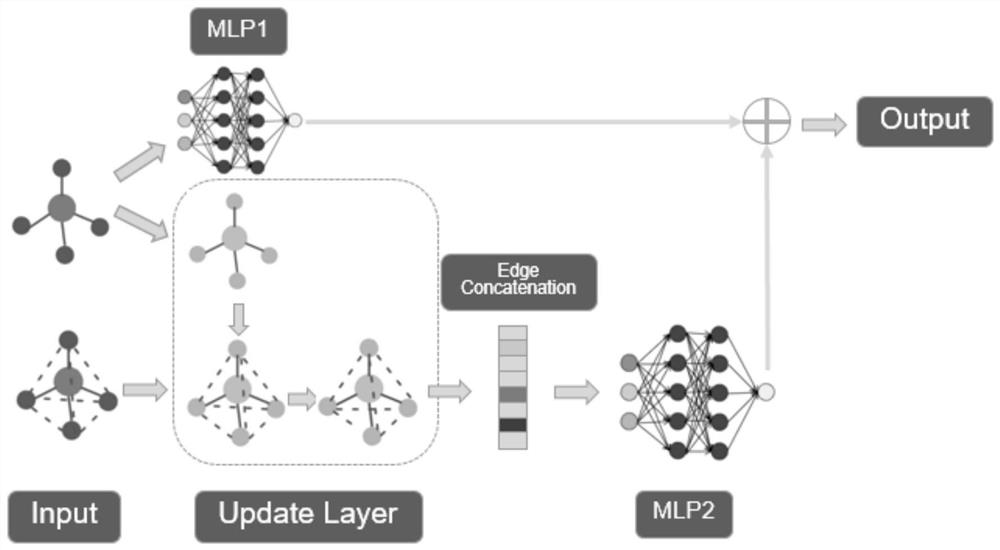
[0019] The present invention constructs a graph according to the SMILES of the input molecule. The graph is composed of atoms as nodes, bonds as edges, some properties of atoms, such as atomic radius, valence electron arrangement, etc., as feature embeddings of nodes, and bond types as edges Feature embedding. In addition, since the distance matrix of molecules needs to be predicted, when constructing the input graph, the construction of edges between atoms without bonds is also required, that is, to construct a complete graph as the input of the model, and the atomic distance matrix Predictions translate to edge length predictions. The overall framework of the model is shown in figure 1 As shown, since the complete graph destroys the structural information of the original molecular graph, the model sets up two branches to process the complete graph and the m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com