Pseudomonas plecoglossicida aptamers and screening method therefor
A Pseudomonas mutans, screening method technology, applied in biochemical equipment and methods, DNA preparation, DNA / RNA fragments, etc., can solve problems such as inability to fully explain pathogenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
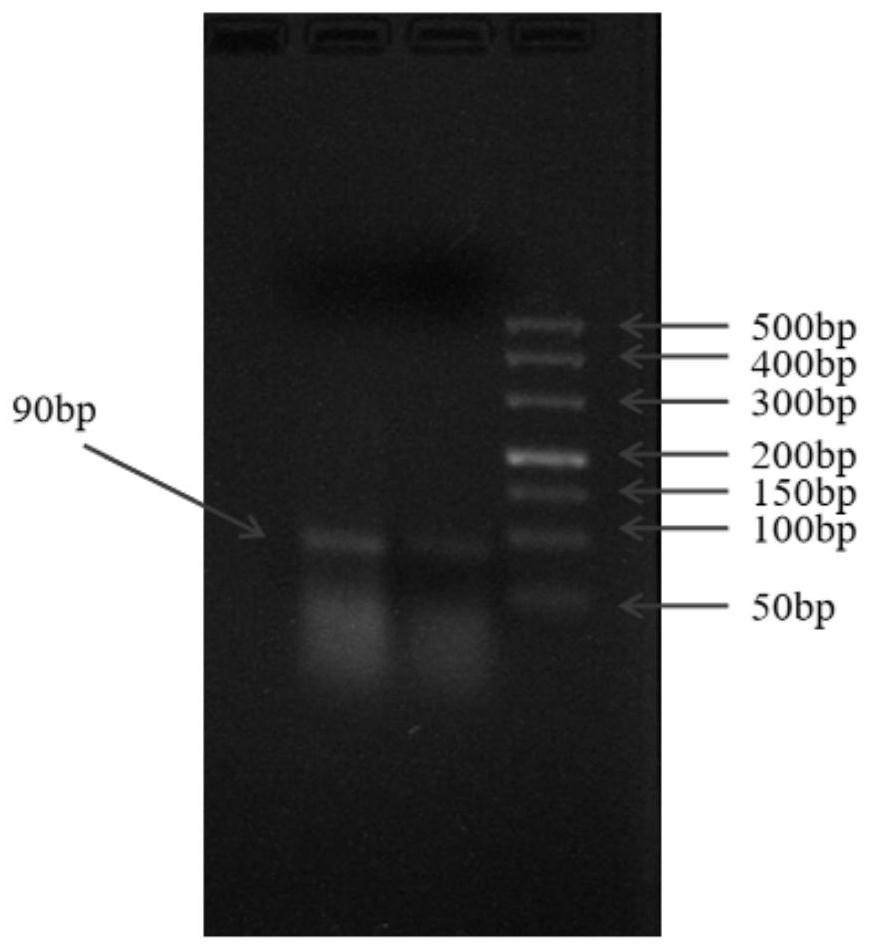
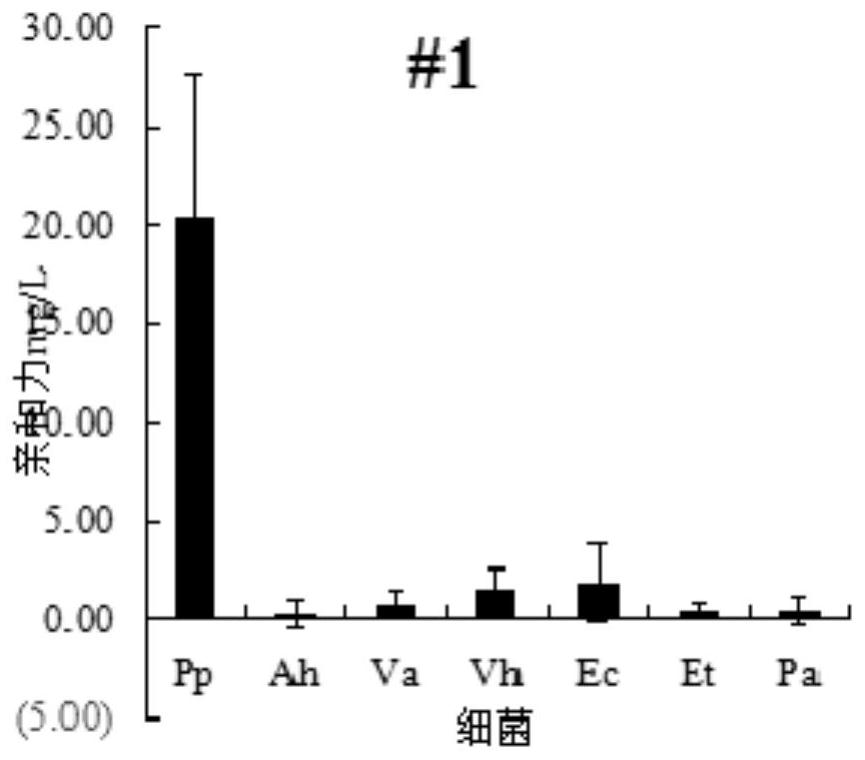
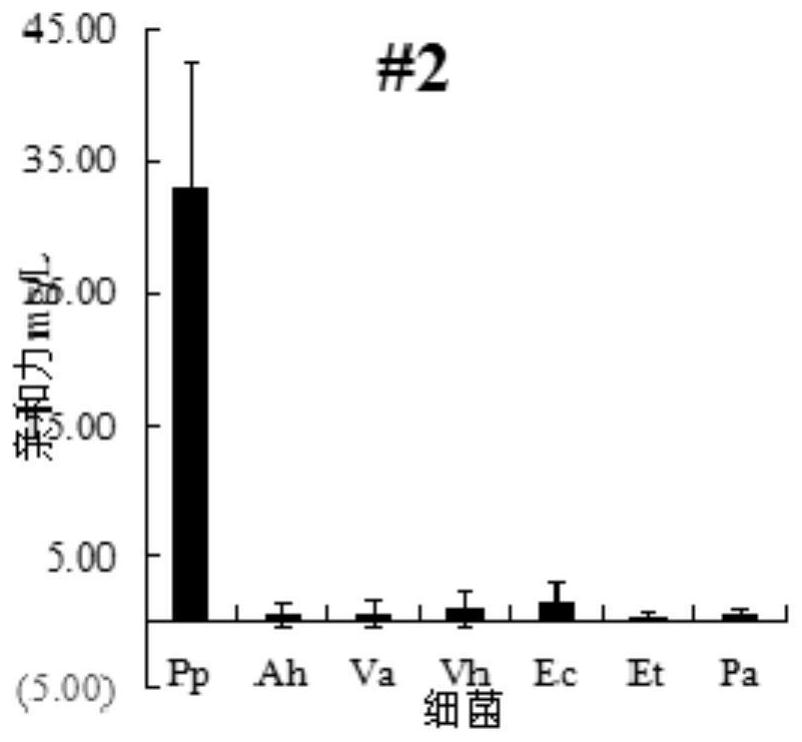
[0036] The present invention takes Pseudomonas mutans as an object, adopts reverse screening and other SELEX technologies to screen nucleic acid aptamers that can specifically recognize Pseudomonas mutans, and then verifies the affinity of the nucleic acid aptamers.
[0037] 1 SELEX screening of nucleic acid aptamers
[0038] (1) Bacteria treatment: take the cultured Pseudomonas mutans centrifuge tube, centrifuge at 6000r / min for 5min, discard the supernatant, wash the bacteria 3 times, add sterile culture medium and mix evenly, measure the OD value of the bacteria solution, Then take the bacteria content of about 4 × 10 8 Centrifuge the Pseudomonas mutans solution at 6000r / min for 5min, discard the supernatant, wash the bacterial pellet three times with 0.9% normal saline, wash the bacterial pellet once with 1×binding buffer, and finally add 100μL of 2×binding buffer Resuspended.
[0039] (2) Binding: take ssDNA random library, dilute to 2μmol / L100μL with 2×binding buffer, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com