Aptamer specifically binding to reverse transcriptase and application thereof
A nucleic acid aptamer and reverse transcriptase technology, which can be used in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve problems such as reverse transcriptase enzyme antibody inactivation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Screening of nucleic acid aptamers specifically binding to reverse transcriptase
[0028] 1. ssDNA random library and enrichment library:
[0029] The DNA random library and primer sequences used were synthesized by Sangon Bioengineering (Shanghai, China) Co., Ltd. and purified by High Performance Liquid Chromatography (HPLC).
[0030] 2. Screening process of nucleic acid aptamers
[0031] The 200pm random ssDNA library was denatured in Hank’s solution at 95°C for 5 minutes, and ice-bathed for 10 minutes. In order to reduce non-specific binding, 0.1 mg / ml salmonspermDNA and 1 mg / ml BSA were added to Hank’s solution as binding buffer. Then, the random library and the magnetic beads coupled with 2ug target polypeptide were reacted in 200ulbinding buffer at 37°C for 30min, and Hank’s washed 3 times. Finally, the magnetic beads bound with ssDNA were used as the template, and the dsDNA was amplified with primers labeled with FITC and biotin. Double-stranded prod...
Embodiment 2
[0049] The using method of embodiment 2 reverse transcriptase nucleic acid aptamers
[0050] 1. Reaction system
[0051] In this embodiment, the reverse transcriptase nucleic acid aptamer of the present invention is used in the following reaction system, which includes reagent R1 and reagent R2 that are independent of each other.
[0052] (1) Reagent R1 is prepared according to the following formula:
[0053] components Serving per person (μL) unit / concentration 10×Buffer for Taq 3.2 / MgCl2 0.2 100mM dATP 0.01 100mM dCTP 0.01 100mM dGTP 0.01 100mM dUTP 0.02 100mM h 2 o
3.54 /
[0054] (2) Reagent R2 is prepared according to the following formula:
[0055] Enzyme composition Per serving (µl) unit / concentration Taq 0.3 5U / μL Anti-Taq 0.3 5U / μL RNase Inhibitor 0.05 40U / μL UNG, heat-labile 0.02 2U / μL RT 0.005 40U / μL aptamer 0.01 1OD / mL h ...
Embodiment 3
[0061] Example 3 Performance Verification of Reverse Transcriptase Nucleic Aptamer
[0062] A total of 3 sets of experiments were set up, and the difference between the kit used in each set of experiments and Example 2 was only the reverse transcriptase nucleic acid aptamer in reagent R2:
[0063] Group A: select the nucleic acid aptamer with the sequence shown in SEQ ID NO.1 to prepare a PCR reaction system;
[0064] Group B: no reverse transcriptase nucleic acid aptamer was added, and 0.01 μL ultrapure water was added at the same time.
[0065] (1) Application of reverse transcriptase nucleic acid aptamer in PCR warm start:
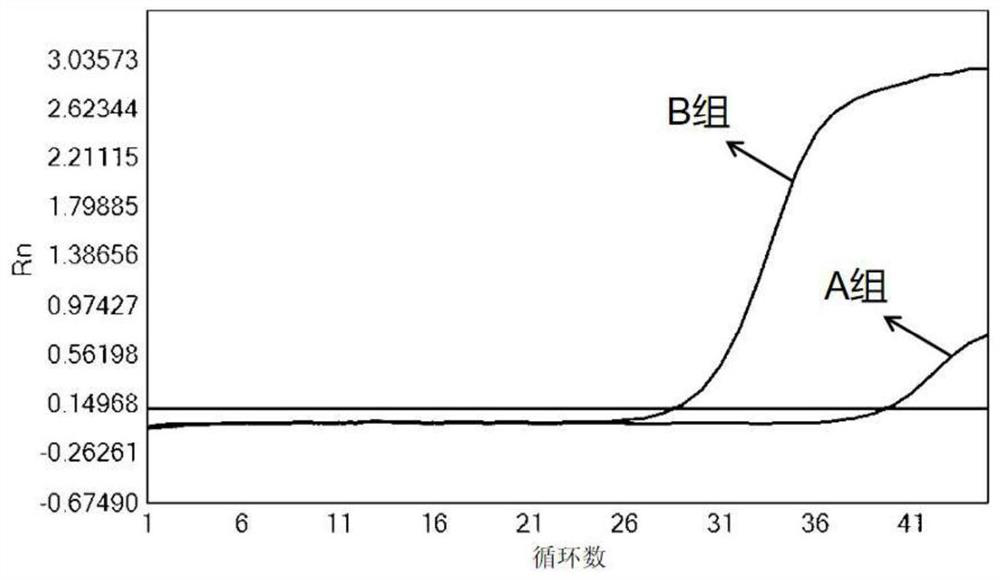
[0066] Using ultrapure water as a template, the PCR reaction was carried out according to the reaction procedure described in Example 2. The result of the reaction is as figure 1 As shown, the results show that: in the reaction system without template, the CT value of group B is significantly lower than that of group A, indicating that more primer di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com