Nucleic acid detection method based on PCR amplification and CRISPR-Cas12a and application
An amplification reaction, nucleic acid technology, applied in the fields of molecular biology and microbiology, can solve the problems of long time required for isolation and culture, misdiagnosis, delayed treatment and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1. Construction of CRISPR-Pa method
[0032] The workflow of CRISPR-Pa disclosed by the present invention:
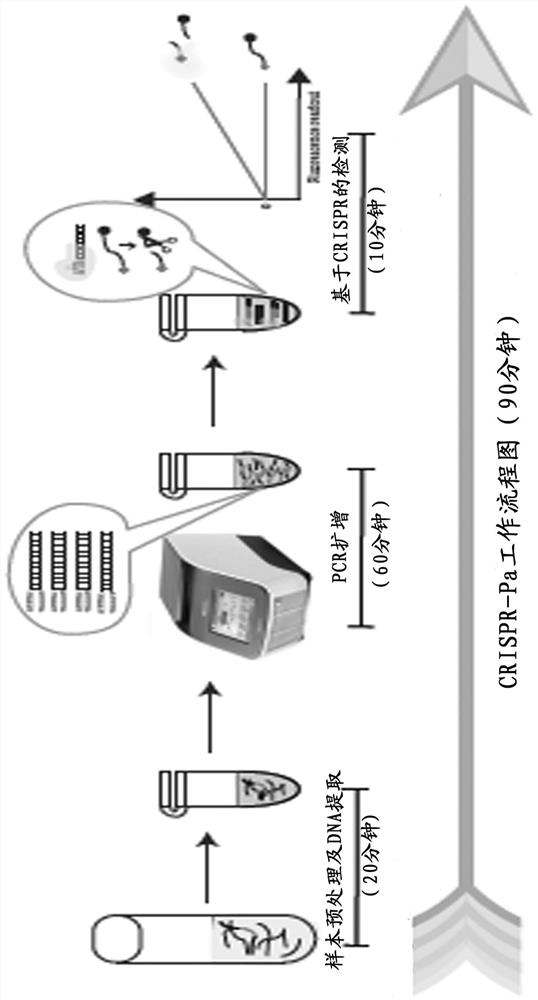
[0033] Such as figure 1 As shown, the CRISPR-Pa platform includes the following three steps: 1. Sample pretreatment and DNA extraction, about 20 minutes; 2. PCR amplification, 60 minutes; 3. CRISPR-based fluorescence detection, 10 minutes. The entire CRISPR-Pa detection process can be completed within 90 minutes.
[0034] The detection principle of CRISPR-Pa disclosed in the present invention:
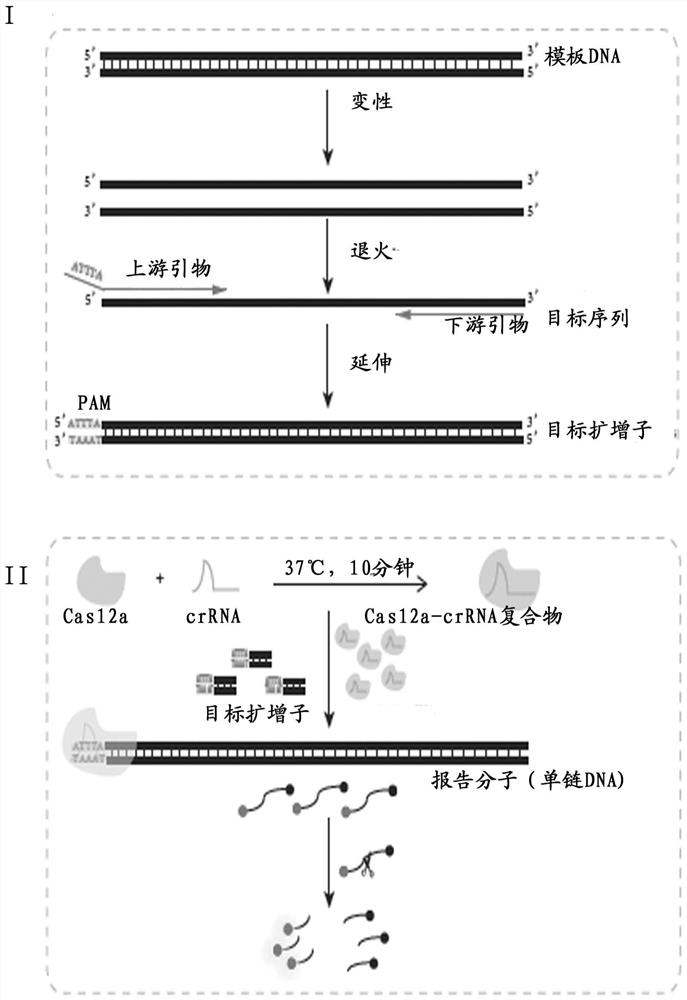
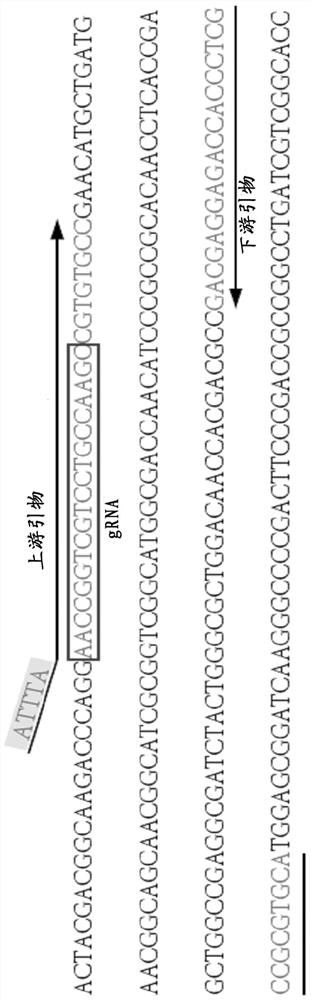
[0035] Such as figure 2 As shown, the detection process after DNA extraction is divided into two steps: 1. The target sequence is exponentially amplified by PCR. In the annealing stage of PCR, the upstream and downstream primers modified with the PAM site: ATTTA (SEQ ID NO: 1) bind to the template DNA strand, and are copied together with the template DNA strand in the amplification stage. After PCR amplification, the upstream of the target amplicon carrie...
Embodiment 2
[0062] Example 2. Sensitivity evaluation of Nocardia melioides CRISPR-Pa detection system
[0063] CRISPR-Pa was performed using the genomic DNA of the standard strain IFM 10152 of Nocardia melioli, which was serially diluted, and the results were as follows: Figure 5 show, Figure 5 Among them, 1-9 are 10ng, 1ng, 100pg, 10pg, 1pg, 100fg, 10fg, 1fg template DNA per reaction and blank control respectively. The lower limit of detection of Nocardia mallei CRISPR-Pa detection system for pure culture bacteria is 1 pg of template DNA.
Embodiment 3
[0064] Example 3. Optimization of the reaction conditions of the Nocardia melioides CRISPR-Pa detection system
[0065] Under standard reaction system conditions, the PCR amplification annealing temperature is carried out at 61-73° C. (2° C. increment). After PCR amplification, 2% agarose gel electrophoresis was used to detect and evaluate the amount of PCR amplification products at different annealing temperatures, and CRISPR detection was used to further evaluate the detection results. Such as Image 6 As shown, positive amplification can occur when the standard reaction system is annealed at 61-73°C, and 69-73°C is the best. In CRISPR assays, such as Figure 7 As shown, all positive results appeared within 10 minutes of real-time fluorescence signal detection, and the fluorescence value of the PCR product annealed at 69°C was the highest. Therefore, we believe that the optimal reaction time for N. mallei CRISPR-Pa detection at 37°C is 10 minutes, and the optimal annealin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com